Originally published : Mon, March 27, 2023 @ 3:59 PM
Updated : Mon, June 26, 2023 @ 10:24 AM
Oligonucleotide therapeutics have unlocked new treatment possibilities for rare and fatal diseases often associated with undruggable proteins.1 Here, synthesised oligonucleotides are used to modulate gene expression by inhibiting, replacing, adding, or editing RNA or DNA in the target cells. Some of the major types of oligo therapeutics currently under development include small interfering RNA (siRNA), antisense oligonucleotides (ASOs), aptamers and CRISPR.1,2 Additionally, oligonucleotides can be tailored to generate patient-specific sequences, making them a favourable tool for precision and personalised medicine.
As the future of oligonucleotide therapies holds great promise, you may be considering in-house oligo synthesis. This strategy helps gain greater control over the production process, and brings benefits such as improving turnaround time for synthesis and safeguarding intellectual property.
Given the various oligonucleotide therapeutic approaches and nucleotide modifications available, choosing the right solid support is essential for the synthesis of therapeutic-grade oligos. Read on for the basics on controlled pore glass (CPG) solid supports and to understand how to select the right support for your specific application.
Controlled pore glass for solid-phase oligo synthesis
 Oligonucleotide synthesis is typically carried out using the phosphoramidite method on a solid support, a process known as solid-phase synthesis. This involves attaching the starting nucleotide (3' end) to a solid surface using a linker, and then adding subsequent nucleotides to the chain in the 3' to 5' direction. CPG matrix has emerged as the preferred support material for oligo synthesis, owing to its superior properties when compared with materials such as cellulose and silica gel. Derived from glass, they are non-swellable, chemically inert, and produce less backpressure when packed into continuous-flow columns.3 CPGs of pore sizes ranging from 500 to 3000 Å are commonly used in oligonucleotide synthesis, with smaller pore sizes allowing for higher nucleoside loading (figure 1).
Oligonucleotide synthesis is typically carried out using the phosphoramidite method on a solid support, a process known as solid-phase synthesis. This involves attaching the starting nucleotide (3' end) to a solid surface using a linker, and then adding subsequent nucleotides to the chain in the 3' to 5' direction. CPG matrix has emerged as the preferred support material for oligo synthesis, owing to its superior properties when compared with materials such as cellulose and silica gel. Derived from glass, they are non-swellable, chemically inert, and produce less backpressure when packed into continuous-flow columns.3 CPGs of pore sizes ranging from 500 to 3000 Å are commonly used in oligonucleotide synthesis, with smaller pore sizes allowing for higher nucleoside loading (figure 1).
The standard practice involves loading CPG columns into an automated synthesizer for improved efficiency and accuracy. The resin is usually 125 to 177 µm in diameter, and derivatised with a terminal nucleotide attached to a long spacer arm.4
Selecting the right CPG solid support for therapeutic applications
The CPG particle size and shape, pore size, pore volume and specific surface area can directly influence the solution exchange behaviour, ligand loading and distribution and reaction kinetics, affecting the efficiency, purity, and reproducibility of oligo synthesis.
The main difference between oligo synthesis for therapeutics and diagnostics or research is the scale. For diagnostic and research purposes, oligos are typically synthesized at a small to medium scale where higher pore sizes are more appropriate. Whereas therapeutic applications require a larger scale and necessitates the use of highly loaded 500/600 Å CPGs.
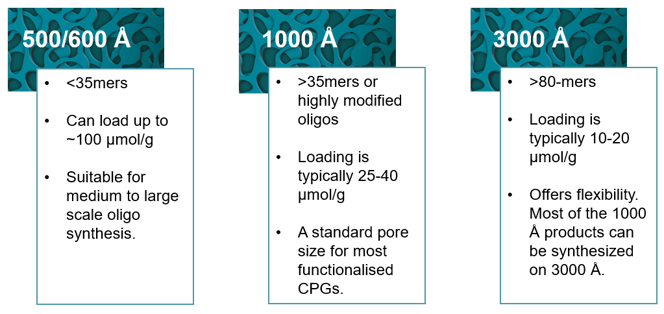
Figure 1: Pore size selection guide. The choice of pore size required depends on the length, complexity, and intended use of the oligonucleotide. Longer oligonucleotides and highly modified oligonucleotides may require larger pore sizes to prevent blockage and improve yields.
CPG modifications
For standard unmodified oligos, a solid support is used that has the 3' base attached to it. Universal supports do not have any base attached to it and can be used to initiate any oligo synthesis. However, CPG supports can be functionalised with suitable modifiers, appropriate for the intended application. With respect to therapeutic oligos, such as ASOs and siRNAs, modifiers are often incorporated at the 3′ end to improve in vivo stability, cell delivery and uptake.
Keeping these factors in mind, we'll consider which CPG is appropriate for your application.
If you're working with RNA-interference technologies
Unmodified oligonucleotides have a short half-life in circulation and cannot permeate intact cellular membranes,5 necessitating conjugation for improved stability, bioavailability, and circulation time. Modified CPGs are used for siRNA or anti-miRNA synthesis based on the desired conjugation approach.
Need a cell delivery modification?
To deliver siRNAs to target cells and improve uptake, oligonucleotides are conjugated with lipids or cell-specific molecules at the 3′ or 5′ end. Here are the specialised CPG supports for incorporating cell-delivery modifiers into 3′ end of your oligo drug molecules:
GalNAc
N-acetylgalactosamine (GalNAc) conjugates act as ligands for the asialoglycoprotein receptor (ASGPR) on hepatic cells, enabling targeted drug delivery to the liver. Compared to unmodified antisense oligonucleotides, GalNAc conjugation increases hepatocyte delivery by ~10 times and is already used in four approved siRNA therapeutics.6
dR-GalNAc (Alpha) CPG by LGC Biosearch Technologies with a dR spacer, can be used to add GalNAc residues to the 3' end of an oligonucleotide. They are available as bulk or columns in pore size variants.
Cholesterol
Lipophilic conjugates have proven effective in delivering single- and double-stranded RNAs, including ASOs, siRNAs, Dicer substrates, and asymmetric siRNAs.5 Cholesterol conjugation promotes oligonucleotide uptake through two mechanisms:
- The cholesterol conjugate intercalates into the plasma membrane, facilitating internalisation via endocytosis.
- It binds to circulating plasma lipoproteins and promotes uptake through interactions with lipoprotein receptors.
Additionally, conjugation of siRNA to cholesterol has demonstrated efficient and consistent loading of extracellular vesicles with the therapeutic cargo.7
Biosearch Technologies offers various CPG supports modified with plant-derived cholesterol, ensuring regulatory compliance. They are available in varying pore sizes and functional loading capacities in bulk and column formats, for the incorporation of cholesterol at the 3' end of an oligonucleotide.
Palmitate
Palmitoyl, a lipid moiety, is similar to cholesterol in its ability to increase the hydrophobicity of oligonucleotides and enhance their cellular uptake. This strategy has been used to modify a thio-phosphoramidate oligonucleotide, GRN163, resulting in improved telomerase inhibition potency.8
Learn more about the 3'-Palmitate CPG (bulk or column) from Biosearch Technologies.
Please note that we don’t currently have a solid support that truly fits an ASO application, but we'll be happy to develop a customised solution if you get in touch and tell us about your requirements.

No need for a cell delivery modification?
If you do not require any 3′ end modification, unmodified CPGs may be the best option. The 500/600 CPGs support large scale oligo synthesis and can load up to ~100 μmol/g.
However, unmodified oligos are susceptible to exo- and endo-nucleases, impacting their therapeutic efficiency. Hence, introducing nuclease resistance is highly recommended. CPG supports that confer nuclease resistant modifications at 3′ are discussed below:
2′-O-Methylation
2'-O-methylation is a chemical modification at the 2′ position of the sugar ring that offers intermediate nuclease resistance between unmodified nucleosides and the highly resistant phosphorothiolation.9 When a high level of nuclease resistance is required, extensive 2'-O-methylation is a popular choice. Moreover, 2'-O-methylation enhances the binding affinity of oligos to their target, resulting in a higher duplex Tm. For these reasons, 2'-OMe nucleosides are extensively used in siRNA and aptamer applications. Additionally, siRNAs modified with 2′-O-Methyl guanosine and uridine residues were found to decrease the immunostimulation and interferon response, improving therapeutic efficacy.10
Functionalised CPG supports can be used to directly incorporate 2′-OMe modified nucleotides at the 3′ end of your oligonucleotide. We offer a range of 2'-OMe CPGs with different pore sizes and linkers that are consistent with our unmodified DNA and RNA CPG products.
2′-Fluoro
2'-F-RNA oligonucleotides have a higher affinity to RNA targets and adopt an A-form helix upon hybridisation, rendering them resistant to nucleases. As a result, aptamers and siRNAs composed of 2'-F-RNA show increased stability in vivo.11,12 Moreover, it could potentially reduce the cytotoxicity associated with the use of ASO or siRNA. A study demonstrated that the introduction of 5-fluoro-2'-deoxyuridine (FdU) moieties at multiple positions of siRNA targeting thymidylate synthase (TS) resulted in a 10-100 fold improvement in cytotoxicity compared to unmodified siRNA.13
We provide a range of 2'-F CPGs for the incorporation of a 2'-fluoro-modified nucleobase at the 3' end of an oligonucleotide. They are available in a variety of pore sizes and linkers consistent with our unmodified DNA and RNA CPG products.
If you're working on CRISPR applications
Despite being a relatively new field, CRISPR-based therapeutics have been gaining momentum and intense research is being conducted on ex vivo and in vivo gene editing across various therapeutic areas.14 Modifying the guide RNA (gRNA) is crucial for improving the CRISPR-Cas9 system's flexibility and efficacy as a genome-editing tool. By engineering gRNAs, the CRISPR system's stability, specificity, safety, and versatility can be enhanced. These modifications aim to protect against nuclease degradation, improve affinity to reduce off-target editing and minimise cytotoxicity.15
Recent research has shown that incorporating 2'-O-methyl modifications at the 5' and 3' ends of the gRNA increases stability, resulting in a higher frequency of indels at the on-target site compared to unmodified sgRNA.15 Additionally, the inclusion of 2'-O-methyl and phosphorothioate groups on the 2'-OH and phosphate backbone within synthesised gRNAs has been found to completely eliminate any immune response.16
To optimise the synthesis of CRISPR gRNA, the choice of functionalised CPG supports depends on several factors, including the desired modification, scale of synthesis, and length of gRNA. For best results, we suggest selecting CPG supports with large pores, 2000/3000 Å, as they offer flexibility and are suited for CRISPR applications.
Maximising the potential of oligo therapeutics with customised CPG columns
Our CPG supports are optimised for particle size and shape, pore size, pore volume, and specific surface area to improve the purity and yield of therapeutic-grade oligonucleotides. Explore our range of bestselling CPG supports, suited for the production of the latest therapeutic oligo classes including LNA, delivery enhancing lipid ligands, siRNA and single-guide RNA (sgRNA) for CRISPR applications.
More information and technical assistance
Related blogs:
Oligonucleotide synthesis basics: introduction to phosphoramidite chemistry
Nucleic acid therapeutics for next generation disease management
Overcoming manufacturing challenges associated with nucleic acid therapeutics
References:
- Thakur S, Sinhari A, Jain P, & Jadhav HR. (2022). A perspective on oligonucleotide therapy: Approaches to patient customization. Frontiers in Pharmacology, 13. DOI:10.3389/fphar.2022.1006304.
- Takakura K, Kawamura A, Torisu Y, Koido S, Yahagi N, & Saruta M. (2019). The Clinical Potential of Oligonucleotide Therapeutics against Pancreatic Cancer. International Journal of Molecular Sciences, 20(13). DOI:10.3390/ijms20133331.
- Pon RT. (1993). Solid-Phase Supports for Oligonucleotide Synthesis. In: Agrawal, S. (eds) Protocols for Oligonucleotides and Analogs. Methods in Molecular Biology, vol 20. Humana Press. DOI:10.1385/0-89603-281-7:465.
- Ellington E & Pollard Jr JD. (2000). Current Protocols in Nucleic Acid Chemistry A.3C.1-A.3C.22. https://deepblue.lib.umich.edu/bitstream/handle/2027.42/143751/cpnca03c.pdf.
- Osborn MF, Khvorova A. (2018). Improving siRNA Delivery In Vivo Through Lipid Conjugation. Nucleic Acid Ther. 28(3):128-136. DOI:10.1089/nat.2018.0725.
- Prakash TP, Graham MJ, Yu J, et al. (2014). Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 42(13):8796-8807. DOI:10.1093/nar/gku531.
- Haraszti RA, Miller R, Didiot MC, et al. (2018). Optimized Cholesterol-siRNA Chemistry Improves Productive Loading onto Extracellular Vesicles. Mol Ther. 26(8):1973-1982. doi:10.1016/j.ymthe.2018.05.024
- Herbert BS, Gellert GC, Hochreiter A, et al. (2005). Lipid modification of GRN163, an N3'-->P5' thio-phosphoramidate oligonucleotide, enhances the potency of telomerase inhibition. Oncogene. 24(33):5262-5268. doi:10.1038/sj.onc.1208760.
- Monia BP, Johnston JF, Sasmor H, & Cummins LL. (1996). Nuclease Resistance and Antisense Activity of Modified Oligonucleotides Targeted to Ha-ras. J. Biol. Chem. 271, 14533-14540. DOI:10.1074/jbc.271.24.14533.
- Selvam C, Mutisya D, Prakash S, Ranganna K, Thilagavathi R. (2017). Therapeutic potential of chemically modified siRNA: Recent trends. Chem Biol Drug Des. 90(5):665-678. doi:10.1111/cbdd.12993.
- Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, & Cook PD. (1993). Uniformly modified 2'-deoxy-2'-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem., 36, 831-841. DOI:10.1021/jm00059a007.
- Yazbeck DR, Min K-L, Damha MJ. (2002). Molecular requirements for degradation of a modified sense RNA strand by Escherichia coli ribonuclease H1. Nucleic Acids Research, 30, 3015-3025. DOI:10.1093/nar/gkf429.
- Wu SY, Chen TM, Gmeiner WH, Chu E, Schmitz JC. (2013). Development of modified siRNA molecules incorporating 5-fluoro-2'-deoxyuridine residues to enhance cytotoxicity. Nucleic Acids Res. 41(8):4650-4659. doi:10.1093/nar/gkt120.
- Uddin F, Rudin CM, Sen T. (2020). CRISPR Gene Therapy: Applications, Limitations, and Implications for the Future. Front Oncol. 10:1387. DOI:10.3389/fonc.2020.01387.
- Allen D, Rosenberg M, Hendel A. (2021). Using Synthetically Engineered Guide RNAs to Enhance CRISPR Genome Editing Systems in Mammalian Cells. Front Genome Ed. 2:617910. DOI:10.3389/fgeed.2020.617910.
- Schubert MS, Cedrone E, Neun B, Behlke MA, Dobrovolskaia MA. (2018). Chemical modification of CRISPR gRNAs eliminate type I interferon responses in human peripheral blood mononuclear cells. J. Cytokine Biol. 3:121. DOI:10.4172/2576-3881.1000121.


