Originally published : Tue, January 24, 2023 @ 10:56 AM
Updated : Wed, March 22, 2023 @ 1:48 PM
Genotyping plants can be a complex and frustrating process
Imagine you have already decided to use probe-based SNP detection as it has been proven to be reliable and cost-effective in your previous work. You then easily locate your SNPs of interest, and it turns out 80% of them happen to be wedged in the centre of AT-rich or highly repetitive regions. Further to that – you are genotyping sugarcane or strawberries and now have eight sets of chromosomes to deal with. Plus, your samples have only been crudely extracted and contain an abundance of impurities.
Optimising your probes with a Minor Groove Binder (MGB) modification could be your answer.

Let’s get specific – how MGBs can help
With over 40 years’ experience of manufacturing oligos at LGC Biosearch Technologies, we know that for genotyping and SNP detection, making your probes as specific as you can is key.
The most common way of increasing the specificity of your fluorescent probes is to make them as long as possible, but longer probes have a higher propensity for sequence mismatches – an absolute killer for SNP detection. The issue with just shortening your probes, however, is that hybridisation between your probe and target sequence is now hindered and probe binding has also been destabilised.
This is where Minor Groove Binders (MGBs) can help.
The addition of an MGB moiety to your probe increases both its melting temperature (Tm) and specificity,1,2 and at the same time allows you to have a significantly shorter length probe. This is crucial when considering the extremely high frequency of SNPs in some species of plant – for example, it was recently shown that a certain haplotype of potato had a SNP every 29 bp.3 Ideally, we just want to be investigating our SNP of interest and not all the flanking ones.
Another advantage to using shorter probes is the benefit of improved quenching efficiency (the quencher is positioned physically closer to the fluorophore), meaning even lower background fluorescence and better signal-to-noise ratio.
Get into the groove
As the name suggests, Minor Groove Binders (MGBs) are molecules that bind to the minor groove of the DNA helix. Biosearch Technologies’ MGB Probes are fluorogenic probes (typically 8-30 bases in length) with a 3′ non-fluorescent quencher conjugated to a MGB moiety.
MGB modifications improve hybridisation of the probe to your sequence of interest via van der Waals forces (as opposed to covalent or ionic bonding). The addition of MGBs can also significantly improve mismatch discrimination4 – especially when the mismatch is within the MGB region of your probe.
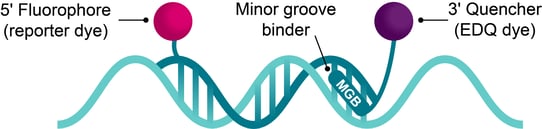
Figure 1: MGB probes are dual-labelled 5′ hydrolysis probes consisting of a 5′ fluorescent reporter dye and a 3′ quencher, conjugated to an MGB moiety.
Crude DNA
The purity and integrity of your extracted DNA is another factor that can heavily influence the calibre of your genotyping data – the phrase ‘you get out what you put in’ springs to mind. However, purifying DNA extracts takes more time and money – and when you are working with high numbers of samples this may not be feasible.
|
|
Because of the issues many breeders and researchers have with crude DNA samples (high quantity of enzyme inhibiting factors), we decided to put our MGB Probes through their paces in combination with our BHQ™ Probe Master Mix. This Master Mix has been thoroughly vetted in conjunction with challenging plant types for SNP identification, so was the logical choice for comparing the results from crude and purified DNA. We found that the combination of MGB Probes and BHQ Probe Master Mix was highly specific and sensitive, even with crude DNA extracts from often difficult plants, like maize and soy – no purification needed! As well as this, both end-point PCR and qPCR applications were tested in volumes of as low as 1 µL, showing that this solution is not just robust, but cost-effective and easy to scale up. |
Plant pathogen detection
The demand for high-throughput assays for the quantitative detection of plant pathogens continues to rise, along with the farmer's desire to only use crop protection chemistry where necessary – we truly are entering the age of ‘Precision Agriculture’. Issues such as environmental pest management, food safety, and global warming are ever increasing worries. Phytopathogenic fungal species, for example, can easily cause a severe drop in both crop quality and overall yields – devastating communities and businesses alike.5
qPCR methods are widely used for the monitoring and identification of a variety of plant pathogens, due to its high specificity. Unfortunately, finding a long enough sequence that is conserved over multiple viral strains (for example), can be an impossible task. Degraded starting material is also common in many workflows, adding further frustration. Trying to diagnose an infection before the visual symptoms set in could be a real challenge too due to minimal quantities of pathogen present in plant samples. Your assay’s sensitivity or Limit of Detection (LoD) will be critical in these situations, meaning that pathogens can remain undetected if the LoD is not low enough.

For this application, MGB probes with their shorter length and improved quenching efficiency can be invaluable. Studies have shown that MGB probe qPCR can be up to 1000x more sensitive than conventional qPCR,6 making your results far more accurate and in some cases the only way to detect a pathogen. Degraded DNA is much less of an issue, MGB probes have even been used to successfully genotype plant varieties used for manufacturing of canned fruit and juice.7 Multiplexing your qPCR assay is also an option,8 giving you the ability to easily distinguish between two pathogens that have the same phenotype in the field.
How probe-based PCR works
Probe-based gene expression analysis and SNP genotyping combine the enzymatic activity of Taq Polymerase, a pair of primers that flank your region of interest, and two dual-labelled probes to amplify and detect your sequence of interest.
- Target dsDNA is denatured by high temperatures, the MGB probe’s fluorophore is emitting no signal as it is positioned in close proximity to the 3′ quencher.
- The temperature is reduced and the PCR primers and sequence specific MGB probe anneal to their target sequences.
- The MGB probe that does not complement the sequence of interest does not anneal to the sequence and remains tethered to its quencher.
- A raised temperature activates Taq polymerase to generate the complimentary DNA strand, and when it reaches the MGB probe it degrades it via 5′ nuclease activity.
- The fluorophore (reporting dye) indicating the SNP/pathogen presence in the sample is now separate from the quencher and fluorescence can now be detected.
This technology can be used in end-point PCR (with the fluorescent signal ratio between the two probes indicating the genotype in the sample) or in qPCR for applications such as GMO testing, Copy Number Variations (CNV), ploidy confirmation and assessment of pathogen load in plant disease diagnostics. As this is a PCR assay, scale can be easily increased by incorporating high-throughput automated platforms, such as the IntelliQube or SNPline/Nexar instruments.
What we can do for you
Biosearch Technologies have licence-free MGB probes available to order, with the choice of six different fluorophores. All our oligos can be customised to your specifications and are manufactured to ISO 13485 or ISO 9001 standards (as required).
We have manufacturing sites both in Europe and the USA – providing manufacturing redundancy, risk mitigation and surge capacity. Further to this, our chemical manufacturing labs ensure that we produce the majority of the raw materials that are critical to our oligo synthesis pipeline.
Our dedicated technical support team are also on hand to help you with any questions on our products and to help you integrate them into your workflow (techsupport@LGCGroup.com).
Related content
Read our blog post on choosing the best qPCR probe chemistry for your assay.
Read more of our agrigenomics content on the blog.
References
- Lukhtanov, E.A., Kutyavin, I.V., Gamper, H.B. and Meyer Jr, R.B., 1995. Oligodeoxynucleotides with conjugated dihydropyrroloindole oligopeptides: Preparation and hybridization properties. Bioconjugate chemistry, 6(4), pp.418-426.
- Kutyavin, I.V., Afonina, I.A., Mills, A., Gorn, V.V., Lukhtanov, E.A., Belousov, E.S., Singer, M.J., Walburger, D.K., Lokhov, S.G., Gall, A.A. and Dempcy, R., 2000. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic acids research, 28(2), pp.655-661.
- Sevestre, F., Facon, M., Wattebled, F. et al. Facilitating gene editing in potato: a Single-Nucleotide Polymorphism (SNP) map of the Solanum tuberosum L. cv. Desiree genome. Sci Rep 10, 2045 (2020). https://doi.org/10.1038/s41598-020-58985-6
- Kutyavin, I., Lokhov, S., Lukhtanov, E. and Reed, M.W. (2003), Chemistry of Minor Groove Binder–Oligonucleotide Conjugates. Current Protocols in Nucleic Acid Chemistry, 13: 8.4.1-8.4.21. https://doi.org/10.1002/0471142700.nc0804s13
- Doehlemann, G., Ökmen, B., Zhu, W. and Sharon, A., 2017. Plant pathogenic fungi. Microbiology spectrum, 5(1), pp.5-1.
- Teng, Q., Shen, W., Yan, D., Yan, L., Li, X., Li, G., Yang, J. and Li, Z., 2015. Development of a TaqMan MGB RT-PCR for the rapid detection of H3 subtype avian influenza virus circulating in China. Journal of virological methods, 217, pp.64-69.
- Endo, T., Fujii, H., Yoshioka, T., Omura, M. and Shimada, T., 2020. TaqMan-MGB SNP genotyping assay to identify 48 citrus cultivars distributed in the Japanese market. Breeding science, 70(3), pp.363-372.
- Luchi, N., Pepori, A.L., Bartolini, P. et al. Duplex real-time PCR assay for the simultaneous detection of Caliciopsis pinea and Fusarium circinatum in pine samples. Appl Microbiol Biotechnol 102, 7135–7146 (2018). https://doi.org/10.1007/s00253-018-9184-1