Originally published : Mon, February 19, 2024 @ 8:44 PM
In November 2023, the UK became the first country in the world to approve a gene therapy based on CRISPR-Cas9 technology. This Nobel prize-winning technique has been lauded as the future of medicine, and we are now seeing the fruition of decades of research.
The new therapy, Casgevy, treats sickle cell disease and beta thalassaemia, both genetic conditions that affect red blood cells. Casgevy precisely edits the genes in a patient’s bone marrow stem cells to produce fully functional haemoglobin. To date, the only cure for these blood conditions has been a bone marrow transplant, a treatment that carries a risk of complications and is only available to a small number of people.
There are multiple CRISPR-based therapeutics in the pipeline, including those that target cancer, diabetes and heart disease. However, designing such therapies requires careful refinement to ensure that the gene edits are precise. This means that the components must be synthesised with reliable purity regardless of the array of modifications included. In this blog post, we look at the key factors to consider when designing and synthesising oligos for CRISPR.
The power of CRISPR
CRISPR was first observed as a form of immune memory in bacteria that helps to remember and destroy viruses. The CRISPR system works by directing endonucleases known as Cas (CRISPR-associated) proteins to target sequences.
Jennifer Doudna and Emmanuelle Charpentier developed CRISPR as a gene editing technology using Cas9 from Streptococcus pyogenes.1 They harnessed this system by designing a guide RNA (gRNA) that directs Cas9 to the exact spot in the genome where a cut is desired. When the gRNA hybridises with the target DNA, it drives conformational changes in the nuclease domains on Cas9, resulting in DNA cleavage. As the cell attempts to repair the DNA break, a template can be introduced to add or replace a sequence, or an imprecise repair can potentially disable the gene through a frameshift mutation.
Charpentier and Doudna showed that two parts of the guide RNA in the bacterial system could be replaced by a synthetic variant, known as single guide RNA (sgRNA), that combines their properties.1 This synthetic format, approximately 100 nucleotides long, gained popularity due to its efficiency and simplicity. 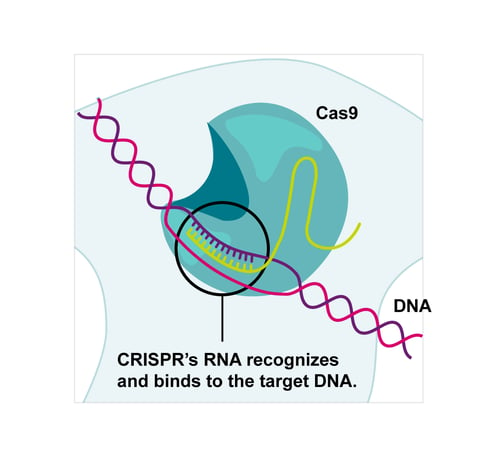
Figure 1: A representation of how the sgRNA guides the Cas9 to cleave the target DNA.
The sgRNA’s critical role means that its structure and stability must be carefully considered to ensure therapeutic impact. However, RNA is inherently unstable and highly susceptible to degradation by endo- and exo-nucleases. This instability poses a challenge for the effective use of sgRNA in clinical settings that can be overcome through a careful approach to its design and production.
Choosing the right controlled pore glass
Controlled Pore Glass (CPG) has been widely used as a solid support for oligo synthesis for several decades. The physical characteristics of CPG, such as particle size and shape, pore size, pore volume, and specific surface area, are critical. These factors directly affect solution exchange behaviour, ligand loading and distribution, and reaction kinetics. Optimising these parameters enhances the synthesis process, increasing efficiency, purity and reproducibility.
The selection of CPG pore size depends on the oligo’s desired length, complexity, and application. For CRISPR, 2000 Å CPG is typically used in solid-phase synthesis of the sgRNA. This pore size is optimal for synthesising longer oligos while maintaining higher yield possibilities, which are characteristic of lower pore sizes. Further, 3000 Å CPG is more suitable for even longer oligonucleotides, such as those exceeding 80 nucleotides.
LGC Biosearch Technologies has perfected CPG manufacture for maximum oligo purity and yield. Our advanced CPG production techniques were developed originally in our Prime Synthesis business, which has produced controlled pore glass since the 1980s.
This expertise has led to carefully refined improvements and consistency in the characteristics of controlled pore glass by Biosearch Technologies.
We have developed proprietary chemical attachment procedures to optimise ligand distributions, providing increased access to reagents and washing solutions and facilitating greater oligo yields and purities. In addition, process refinements and proprietary assays have minimised the troublesome “N-1” impurity levels in an oligo synthesis.
As the demand for oligo therapies continues to grow, we are exploring new manufacturing processes for CPG that are more efficient, reduce environmental impact and are better suited for scaling up. Our new native CPG process maintains high-quality production while increasing manufacturing capacity by 200% and reducing wastewater volume by 50%. In addition, optimising our current CPG for various therapeutic applications has shown a 19% increase in productivity in initial proof of concept studies.
In oligo synthesis, the 3' end is determined by the base or modifier functionalisation of the controlled pore glass solid support. Having a range of functionalised 2000 Å and 3000 Å CPGs offers the opportunity to easily tailor the sgRNA.
Modifying sgRNA for better efficacy
Synthetic oligonucleotides, including sgRNA, face a major challenge within cellular environments due to their susceptibility to degradation. This leads to a rapid reduction in the oligonucleotides’ half-life, largely driven by endo- and exo-nuclease enzymes as well as RNA’s inherent chemical instability.
There are several modifications that can make the sgRNA more resistant to nucleases, as well as improve its stability and efficacy. This enhances the applicability of CRISPR-Cas9 as a therapeutic by reducing off-target effects and cytotoxicity.
This protection can be achieved by modifying the ends of the sgRNA, enhancing its stability against exonuclease activity.2 CPG supports can be functionalised with suitable modifiers to incorporate them at the 3′ end to improve the in vivo stability of the sgRNA.
In addition, modification of internal residues of the guide RNA can improve the specificity of hybridisation with the target sequence. Here are some examples of the modifications that can be introduced to sgRNA to enhance performance.
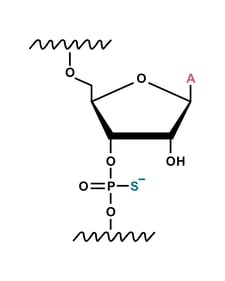 Phosphorothioate
Phosphorothioate
Phosphorothioate modification is a process that enhances the stability and functionality of guide RNA.3 This modification involves replacing the phosphodiester bond, which links sugar moieties in adjacent nucleotides, with a phosphorothioate bond. This substitution is achieved using a sulphurising reagent, which replaces an oxygen atom with a sulphur atom. This significantly stabilises these molecules by reducing their degradation from both extracellular and intracellular nucleases.
We offer DDTT and EDITH sulphurising reagents to efficiently create phosphorothioate linkages in the gRNA backbone.
2'-O-methylation (2'-Ome) modification
The 2'-O-methylation modification, which occurs at the 2' position of the sugar ring, provides less nuclease resistance than phosphorothiolation. So, extensive 2'-O-methylation is needed to ensure a high level of nuclease resistance.
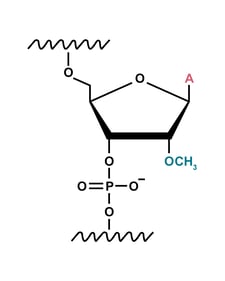 However, it is important not to interrupt the interaction between the sgRNA and Cas9. When all 2′-OH groups are modified, the CRISPR system does not function.4 Using the structure of the complex or a trial-and-error approach can lead to the optimum pattern of modifications to maintain the sgRNA-Cas9 hydrogen bonding without losing nuclease resistance.
However, it is important not to interrupt the interaction between the sgRNA and Cas9. When all 2′-OH groups are modified, the CRISPR system does not function.4 Using the structure of the complex or a trial-and-error approach can lead to the optimum pattern of modifications to maintain the sgRNA-Cas9 hydrogen bonding without losing nuclease resistance.
Adding 2'-O-methyl (2'-OMe) modifications to both the 5' and 3' ends of sgRNA enhances its stability. This stability boost leads to a higher occurrence of insertions and deletions (indels) at the target site, a significant improvement over unmodified sgRNA.
Our range of 2'-OMe CPG supports can directly incorporate 2′-OMe modified nucleotides at the 3′ end of the gRNA. These are available with different pore sizes and linkers suitable for CRISPR gRNA applications.
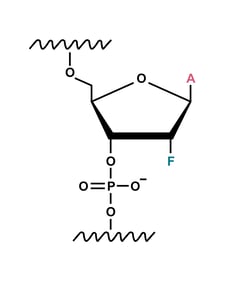 2'-fluoro (2' F) modification
2'-fluoro (2' F) modification
A 2′-fluoro modification increases RNA affinity to DNA. This modification is less bulky than other options, making it less sterically disruptive to Cas9 binding.5
The highly electronegative fluorine replaces the hydroxyl group, a hydrogen bond donor, affecting the conformational preference of the ring. On hybridising to a target, 2′-F-RNA oligonucleotides adopt the more thermodynamically stable A-form helix, increasing the stability of the duplex.6
We provide a variety of pore sizes and linkers for 2'-F CPGs for incorporating a 2'-fluoro-modified nucleotide at the 3' end of gRNA.
Locked nucleic acids (LNAs)
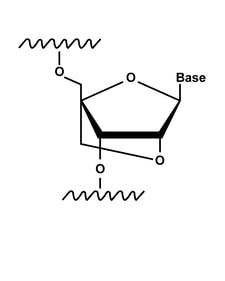 Research has shown that locked nucleic acids (LNAs) and bridged nucleic acids (BNAs) can improve stability and binding efficiency.7,8 These RNA nucleotides are conformationally restricted by linking the 2′ oxygen and the 4′ carbon of the ribose ring.
Research has shown that locked nucleic acids (LNAs) and bridged nucleic acids (BNAs) can improve stability and binding efficiency.7,8 These RNA nucleotides are conformationally restricted by linking the 2′ oxygen and the 4′ carbon of the ribose ring.
In LNAs, the 2′ oxygen and 4′ carbon are directly bonded, while in BNAs, they are linked by a methyl-bound nitrogen. The conformation forced by these modified nucleotides results in increased affinity for complementary sequences and improved mismatch discrimination.
We offer a range of CPGs functionalised with LNAs for introducing these modified nucleotides at the 3' end of gRNA.
A multitude of CPGs for CRISPR applications
Selecting the right controlled pore glass for sgRNA synthesis is crucial to ensure the right purity and yield for therapeutic-grade oligonucleotides. A range of modifications can improve the stability and specificity of the sgRNA, but optimising the combination takes trial and error. If you choose to manufacture the sgRNA yourself, functionalised CPGs give you complete control over introducing 3' modifications.
Related content
- Which controlled pore glass is right for your therapeutic application?
- Know your oligo mod: phosphorothioate bonds
References
- Jinek M et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
- Hendel A et al. (2015) Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 33, 985–989. doi: 10.1038/nbt.3290
- Rahdar M et al. (2015) Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proc. Natl. Acad. Sci. U.S.A. 112, E7110–E7117. doi: 10.1073/pnas.1520883112
- Yin H et al. (2017) Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 35, 1179–1187. doi: 10.1038/nbt.4005
- O’Reilly D et al. (2019) Extensive CRISPR RNA modification reveals chemical compatibility and structure-activity relationships for Cas9 biochemical activity. Nucleic Acids Res. 47, 546–558. doi: 10.1093/nar/gky1214
- Kawasaki AM et al. (1993) Uniformly modified 2'-deoxy-2'-fluoro phosphorothioate oligonucleotides as nuclease resistant antisense compounds with high affinity and specificity for RNA targets. Journal of Medicinal Chemistry 36: 831-841. doi: 10.1021/jm00059a007
- You Y et al. (2006) Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 34:e60. doi: 10.1093/nar/gkl175
- Cromwell CR et al. (2018) Incorporation of bridged nucleic acids into CRISPR RNAs improves Cas9 endonuclease specificity. Nat Commun. 9: 1448. doi: 10.1038/s41467-018-03927-0


