Originally published : Thu, November 13, 2014 @ 4:08 PM

Updated : Mon, September 19, 2022 @ 2:14 PM
Neuroscience gene expression research can come with many hurdles, especially when working with brain tissue or mixed populations of cultured neurons. Here are just a few obstacles Neuroscientists run into:
- The brain is not made only of neurons! Alongside neurons, brain tissue can contain a variety of cell types such as glial cells and oligodendrocytes.These cell types may act separately or in conjunction with each other to perform a specific function.
- The brain itself is regionally heterogeneous, containing specialized anatomical structures such as the amygdala, hypothalamus, and hippocampus. These regions perform distinct functions such as eliciting the fight or flight response, signaling reproduction, or contributing to memory.
- Even so, delving within each of these brain regions, you will find even more specialization. Certain cell populations express a particular RNA signature. For example, tyrosine hydroxylase (Th) expression in dopaminergic neurons, which is critical to the synthesis of the neurotransmitter dopamine. Cells surrounding Th cells, or interspersed with Th cells do not express Th at all! These are all contributing factors that make neuroscience so incredibly fascinating, yet so cumbersome.
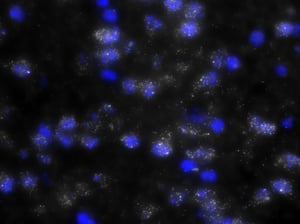
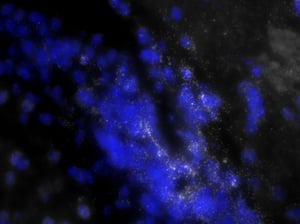
The ability to isolate precise populations in the brain becomes quite a serious challenge. Currently, scientists overcome this by attempting difficult techniques such as laser capture micro-dissection to isolate cells expressing specific genes, or dissociating brain tissue and FACS sorting the cells, or dissecting out whole brain structures to perform qPCR. However, the latter can introduce variability from sample to sample because it is difficult to dissect exactly the same amount of expressing vs. non-expressing cells in one region. Each of these techniques has their advantages and disadvantages. One of the mainstays for Neuroscience research is RNA in situ hybridization (ISH). However, unlike the older ISH techniques, Stellaris RNA FISH enables the simultaneous ability to localize and quantify neurons expressing a gene of interest and their single molecules of RNA.
With many thanks to our ongoing collaboration with the Kauffman lab at UCSD, we recently modified our existing frozen tissue protocol to align with current in situ methods commonly used among Neuroscientists.
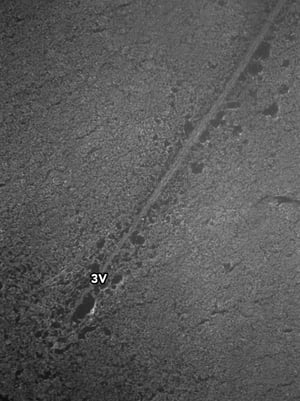
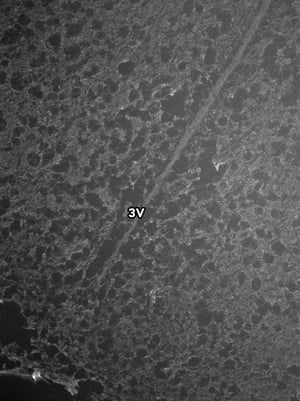
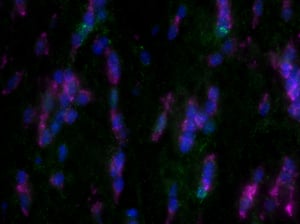
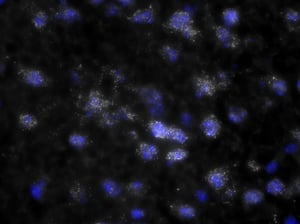
Images were acquired with a 1 second exposure using the FITC filter, which has an excitation of 490/20 nm and emission of 535/45 nm. It is an especially useful filter to gauge autofluorescence in tissue samples. Both images were set to the same display settings, which noticeably highlights the effect TEA and acetic anhydride can have on reducing the inherent autofluorescence in brain tissue. These images were acquired using a 10x objective.
Tissue Preparation
In order to modify our protocols to better suit the Neuroscience field, we utilized frozen mouse brains. These brains were collected, immediately frozen on dry ice, and stored at −80 °C until sectioning. Brains were then cut (coronal) on a cryostat into five series of 10 μm sections, thaw mounted onto Superfrost plus slides, and stored at −80 °C until processing via Stellaris RNA FISH.
Protocol Adjustments
The slide-mounted brain sections were fixed in ice-cold 4% paraformaldehyde for 15 minutes. Following a PBS wash and a few brief equilibration washes, the brain sections were treated for 10 minutes in TEA buffer containing acetic anhydride, which was added fresh, just prior to immersing the slides. Acetylation works to acetylate free amines, such as lysines in proteins contained in the tissue. Presumably, this neutralizes and reduces charge of the tissue, which can diminish unwanted fluorescence. The sections were then rinsed in a 2X sodium citrate, sodium chloride buffer (SSC), followed by a series of ethanol washes at 75, 95, and 100% for 3 minutes each, to dehydrate the tissue. Next, the sections spent 5 minutes immersed in chloroform to de-lipidate the brain tissue, which greatly thins the tissue, making it easier to image. After another brief dehydration in graded ethanols, the tissue sections air dried for 90 minutes prior to the hybridization procedure.
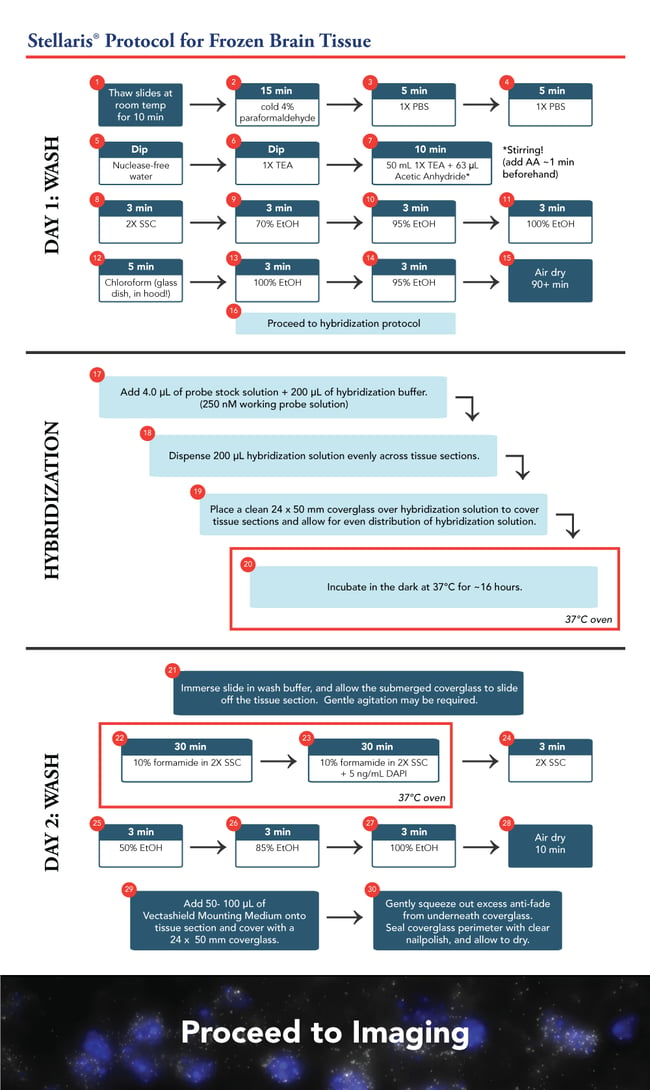
The hybridization procedure remains similar to the current frozen tissue protocol. However, these slides comprised six 10 µm sections of brain tissue each. We scaled up the hybridization concentration and solution accordingly. With our probe at a 12.5 µM stock, we added 4 µL probe +200 µL hybridization buffer to give a 250nM final concentration. We opted for a 24 x 50 mm coverglass rather than an 18 mm square coverglass, to ensure all the tissue sections were covered evenly. The hybridization was processed overnight, approximately 16 hours.
The next morning, the slides were washed twice for 30 minutes using 10% formamide in 2X SSC. This was followed by a brief 2X SSC wash and the tissue sections were dehydrated once more in an ethanol series of 50, 85, and 100%, for 3 minutes each.
As you can see, there is a noticeable decrease in background fluorescence using these protocol modifications. Imaging time was also markedly reduced thanks to the dehydration and de-lipidation steps. They thinned out the tissue so that only 15-22 z-planes were needed for proper imaging (see Page 2 of the Getting Started Guide).
Share Your Feedback
We are continually working to improve our tissue protocol, but we hope these initial improvements will have you performing Stellaris RNA FISH with greater ease in the brain and other tissue types. What are your thoughts regarding the modifications to the tissue protocol? Have you had success? Please feel free to leave a comment, we look forward to hearing from you.