Originally published : Wed, October 20, 2021 @ 9:41 PM
Updated : Fri, June 30, 2023 @ 2:19 PM
Many cultivated crops with agronomic significance, including wheat, peanut, oat, banana, potato, coffee, blueberry and strawberry, are polyploid angiosperms with complex genomes. While polyploidy often generates the potential for more genetic, genomic and phenotypic novelty, linking genotype and phenotype can be more challenging compared to diploid crops.1 The latest genomic technologies are helping plant breeders unravel complex genomes, such as the octoploid strawberry, to improve genetic gain for traits of interest. For example, the latest whole genome sequencing (WGS) technology can increase efficiency, reduce operating costs and improve the data quality for complex plant breeding applications. WGS is an untargeted, next-generation sequencing (NGS) solution and the most comprehensive way to investigate the genome of any species.
What is polyploidy?
Polyploidy is the heritable condition of possessing two or more complex sets of chromosomes.2 Strawberry, the most widely cultivated fruit crop, arose from the hybridisation of two wild octoploid species, formed from the fusion of four diploid subgenomes into a single nucleus more than one million years ago.3
Breeding challenges for polyploid crops
With a complex, highly heterozygous genome, achieving breeding success in polyploids is challenging using traditional breeding methods.4 In diploid crops, only two alleles of the same gene at the same loci on homologous chromosomes contribute to gene expression and the corresponding phenotype. That makes identifying and selecting a trait of interest more straightforward, even if multiple genes contribute to the phenotype.

However, in polyploids, multiple alleles are associated with a single locus, making segregation more complex than diploids. Homeoalleles of a given gene can contribute to complex interactions with a substantial effect on phenotype. For example, in the octoploid strawberry, determining which allele or combination of up to eight different homeoalleles regulates the expression of a trait of interest can be extremely difficult.5 Polyploid plant cells have complex regulatory mechanisms to unify gene expression between the homeologs and define their relative contributions to the final phenotype.1
Marker-assisted selection is one way to improve breeding efficiency for polyploid crops. However, developing and analysing molecular markers and performing sequencing projects can be complicated due to the high sequence homology between subgenomes in polyploids.2
In the past decade, next generation sequencing (NGS) has revolutionised the exploration of polyploid genomes. NGS technologies reduce sequencing costs, increase throughputs and expand fragment assembly complexity due to their short sequence read outputs. A reference genome is used to map and standardise variations across different individuals within a species to aid genetic analysis.1 While applying NGS in polyploids, some possible pitfalls and challenges need to be avoided or taken into consideration, such as uneven and inadequate sequencing depth, copy number uncertainty, heterozygote miscalling, quality of the reference genome, short-read alignment strategy, missing data and sequencing errors.6
Recent research helps overcome breeding challenges in strawberry
As mentioned, a complex octoploid genome has historically challenged genotyping and genetic mapping progress for strawberry breeding. Recent research efforts to address some of the significant technical challenges include:
- the development of a high-quality octoploid genome assembly,
- whole genome sequencing of numerous octoploid individuals to better understand intra- and inter-homoeologous nucleotide variation,
- identification and physical mapping of DNA variants across the octoploid genome and comparative genetic mapping of the wild octoploid progenitors.6
This information helps breeders employ genomic tools to generate more reliable data to inform decision making. Strawberry breeding programs can now use genome-wide prediction for parent selection to improve yield and quality traits. Studies have shown that markers are more effective than pedigrees for estimating breeding values, even when phenotypic information is present.6 In addition, phenotyping efforts can be reduced when selections are made using prediction models built in training populations that were properly phenotyped and genotyped. Another benefit to genomic breeding tools is that individuals with high predicted performance can be used as parents earlier in the breeding cycle, getting products to market sooner.6
Flex-Seq: An efficient, economical solution for genotyping polyploids
Assays for single nucleotide polymorphism (SNP) detection, such as KASP™ polymerase chain reaction (PCR) based genotyping, have become popular for strawberry breeding applications due to an abundance of SNP information from array genotyping, accuracy and ease of scoring and resilience to crude DNA extracts.6 But, major limitations of arrays include the length of time for production and lack of scalability. It can take three to six months to design and manufacture a fixed set of markers, which may not be practical for fast-moving breeding cycles.
LGC Biosearch Technologies’ Flex-Seq™ targeted genotyping solution is a high-throughput and flexible genotyping approach that addresses many critical genotyping challenges that breeders face. Flex-Seq provides a cost-efficient, flexible and scalable mid-plex genotyping platform as a service. With appropriate planning, Flex-Seq process flow enables the design and manufacture of probe libraries, DNA purification, sequencing and data analysis to fit into plant breeding cycles.
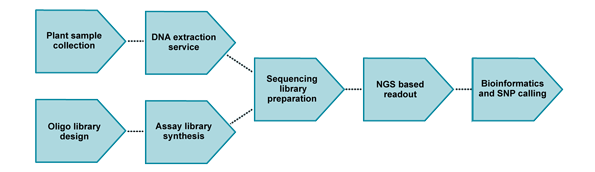
Flex-Seq technology allows complex trait assessment in modern breeding programs and is an ideal option for highly heterotic species or segregating populations in crossing programs. The Flex-Seq probe design offers flexibility for target sequences, avoiding repetitive regions of the genome and is typically not affected by variability in DNA near the target site. Flex-Seq is an excellent solution for genotyping polyploid crops.
Focusing on scalability paired with data accuracy, reproducibility, and completeness allows Flex-Seq to deliver industry-scale solutions for industry-scale genotyping. Flex-Seq genotype data matches chip-based genotyping array technology, ensuring consistency between legacy datasets. Genotyping data from other technologies can also be incorporated into Flex-Seq assays and custom/novel genotyping markers can be developed for any species.
 Flex-Seq’s sister product Capture-Seq™ more broadly targets SNPs, genetic variations within genes and QTLs at the same time to produce contiguous sequence data at each region, instead of a single SNP, through the power of NGS. This enables Capture-Seq to provide greater genomic insights into breeding material and training populations, including the ability to phase alleles into haplotypes for more effective polyploid results.
Flex-Seq’s sister product Capture-Seq™ more broadly targets SNPs, genetic variations within genes and QTLs at the same time to produce contiguous sequence data at each region, instead of a single SNP, through the power of NGS. This enables Capture-Seq to provide greater genomic insights into breeding material and training populations, including the ability to phase alleles into haplotypes for more effective polyploid results.
Comprehensive tools to increase genetic gain
Improving genome sequencing of polyploid crops will have a fundamental impact on genetic research and plant breeding outcomes. With a more thorough understanding of plant genomes, and the ability to efficiently identify genomic variants and tie them to economic, physiological, and morphological agronomic traits, plant breeders will more effectively meet market needs for agronomic crops.1
As an industry leader in the agrigenomics space, Biosearch Technologies has the expertise, experience and products to support complex plant breeding objectives. We offer customised consultations to help recommend the right solutions for each unique plant breeding project. Leaders in the plant breeding industry have vetted our diverse portfolio of products and services.
Watch this webinar to learn more about how Flex-Seq genotyping can help support your breeding objectives.
References
- Kyriakidou M, Tai HH, Anglin NL, et al.Current strategies of polyploid plant genome sequence assembly. Frontiers in Plant Science. 9. Published 2018. Accessed 17 September 2021. https://doi.org/10.3389/fpls.2018.01660
- Woodhouse, M., Burkart-Waco, D. & Comai, L.Nature Education 2(1):1. Published 2009. Accessed 26 January 2023 https://www.nature.com/scitable/topicpage/polyploidy-1552814/#:~:text=Polyploidy%20is%20the%20heritable%20condition,groups%20of%20fish%20and%20amphibians
- Edger PP, Poorten TJ, VanBuren R, et al.Origin and evolution of the octoploid strawberry genome. Nat Genet. 51, 541–547. Published 2019. Accessed 19 September 2021. https://doi.org/10.1038/s41588-019-0356-4
- Schaart JG, van de Wiel CCM and Smulders MJM. Genome editing of polyploid crops: prospects, achievements and bottlenecks. Transgenic Res30, 337–351. Published 2021. Accessed 17 September 2021. https://doi.org/10.1007/s11248-021-00251-0
- Gaston A, Osorio S, Denoyes B, et al. Applying the Solanaceae strategies to strawberry crop improvement. Trends in Plant Science. 25, 2. 130-140. Published 2020. Accessed 17 September 2021. https://doi.org/10.1016/j.tplants.2019.10.003
- Whitaker VM, Knapp SJ, Hardigan MA, et al.A roadmap for research in octoploid strawberry. Hortic Res 7, 33. Published 2020. Accessed 17 September 2021. https://doi.org/10.1038/s41438-020-0252-1


