Originally published : Fri, January 12, 2024 @ 4:37 PM
Creating a crop that is more effective at meeting the needs of farmers and end consumers is the aim of every plant breeder. Using a targeted genotyping by sequencing (GBS) approach can help scientists reduce SNP deserts (regions of the genome with low SNP density) to inform the development of a trait stacking programme, as well as generating the data for germplasm diversity analysis.
The CoverCress® team did just that. CoverCress is a domesticated variant of pennycress, specifically bred to enhance production of oil suitable for renewable fuel and provide protein suitable for animal feed with the potential benefits of cover crops.
It all started in 2013 when three former Monsanto employees collected a large and diverse pennycress collection and created a small company called Arvegenix. Initially, a traditional breeding program focused on yield and maturity was set up. Then in 2016, a parallel effort on the use of gene editing for crop improvement was evaluated and implemented. This combination of traditional breeding and agronomic methods with newer gene editing tools, had brought about the successful launch of a new cash crop for the U.S. Midwest – CoverCress. The crop has two growing cycles – winter and spring – and is well suited to cooler climates.
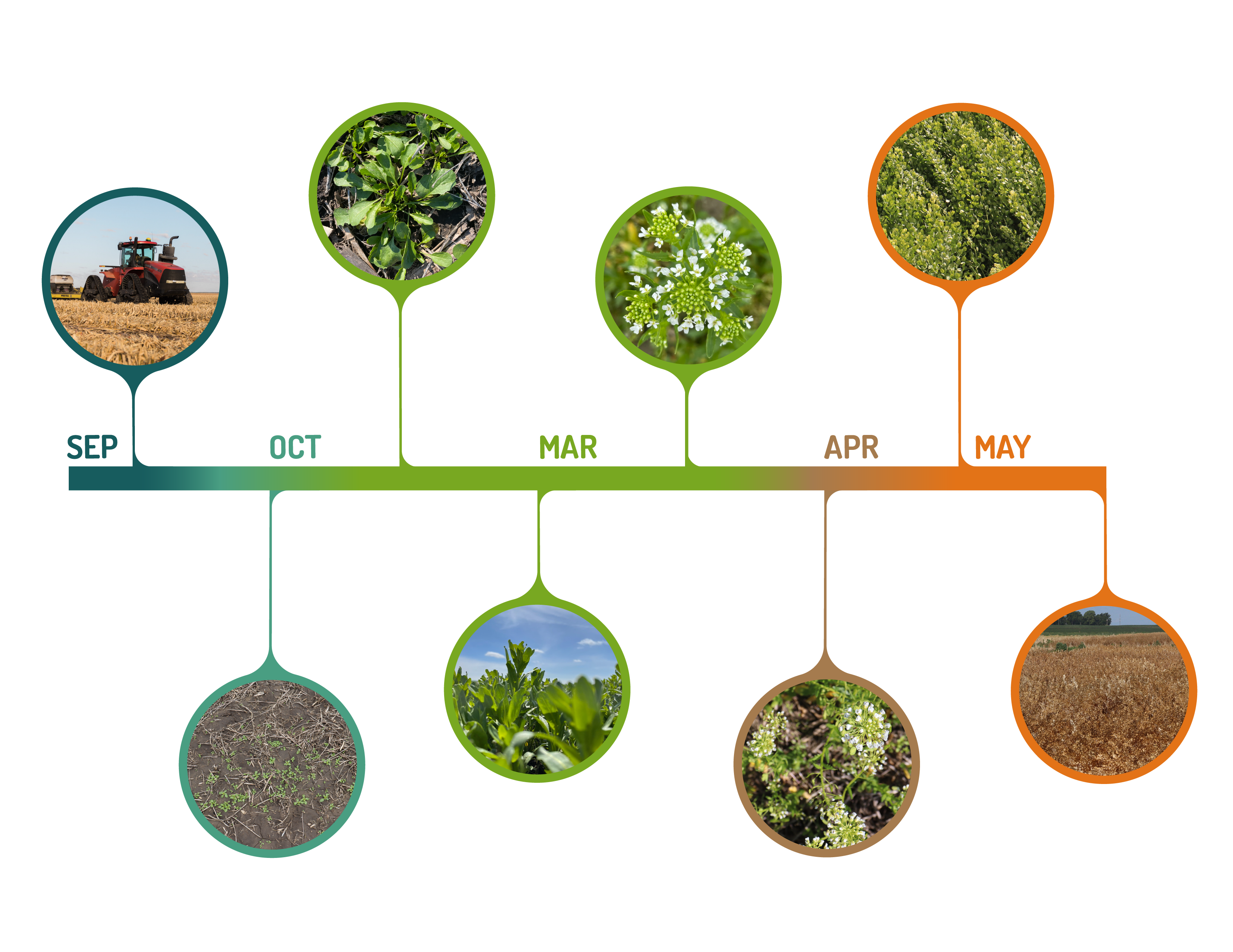 |
| Figure 1. The CoverCress growth cycle |
The Field Pennycress genome
Research on Field Pennycress (Thlaspi arvense) has discovered that it is a self-pollinating diploid with seven chromosomes with a genome size of 500-540MB. Gene space and methylation patterns have also been identified. It is genetically very similar to Arabidopsis and canola, with the gene order highly conserved in many regions.1 This close relatedness facilitated the leveraging of knowledge from Brassica research and the use of gene interaction information to qualify gene candidates for CCI’s product development programme.
Over ten years of research using chemical mutagenesis, gene editing and breeding, CCI has been able to identify primary solutions for fibre content, erucic acid levels, and sinigrin levels as well as other important traits such as pod shatter. As the crop improvement efforts continued, it was evident that traditional genotyping platforms and studies showed limitations and displayed single nucleotide polymorphism (SNP) deserts that potentially impacted the ability to make genomic breeding decisions.
Investigating the genome and why enzyme selection was key
When CCI used whole-genome sequencing (WGS) and then scaled down to targeted resequencing also known as genotyping by sequence (GBS) to inform diversity between CoverCress germplasm with a handful of markers, they found that GBS had limitations in the ability to identify variation across chromosomes. This was due to the methylation patterns found in pennycress which led to SNP deserts that hampered genome-wide assessment causal trait factors. This was disappointing as they wanted to have a cost-effective programme to capture variation across the chromosomes to inform breeding and diversity analysis.
The scientific team at CCI collaborated with scientists at LGC Biosearch Technologies to see if the GBS could be improved. The Biosearch Technologies scientists suggested alternative enzymes and library prep methods to help improve the observations on the frequency of SNPs throughout out the chromosomes and find good marker density. CCI  initially tested 200 genotypes, sequencing them using the suggested enzymes and library prep methods. This pilot study captured 50,000-80,000 variations amongst the germplasm and in doing so they had successfully overcome limitations of GBS and captured the variation required for their germplasm programme.
initially tested 200 genotypes, sequencing them using the suggested enzymes and library prep methods. This pilot study captured 50,000-80,000 variations amongst the germplasm and in doing so they had successfully overcome limitations of GBS and captured the variation required for their germplasm programme.
“Collaboration with the scientists at Biosearch Technologies allowed us to optimise the enzyme combinations that reduced the frequency of SNP deserts and provided good marker density in the population tested”, noted Dr. Ratan Chopra, CCI’s Vice President of Research.
Mapping the genome to inform breeding and trait stacking
However, 50,000-80,000 variations are a lot of markers for breeding and quality control. The next stage of the project was to identify targets that would deliver enough genetic information on the germplasm for the breeding program and quality control.
Dr Chopra and his team had reached a point where a new genotyping platform was required to identify next generation traits, to improve germplasm purity and to enable molecular breeding to accelerate agronomic gains. Impressed with the success of the outcomes of the GBS collaboration with Biosearch Technologies, they chose to work with them again using their targeted genotyping by sequence platform Flex-Seq™, which at the time was very new.
The team at CCI used the GBS data to identify 5,000 regions of interest which captured the genetic variation of CoverCress. They then sequenced these using Flex-Seq, which validated the findings of the GBS and showed the consistency of these markers across the germplasm they had used for GBS. With this validation, they could move to the next stage of the project to create a genetic map of markers that would inform breeding and trait stacking decisions. This was complex, due to the plant’s unique genome which has a bias in gene distribution due to methylation patterns that has limited researchers’ ability to understand genetic linkage.
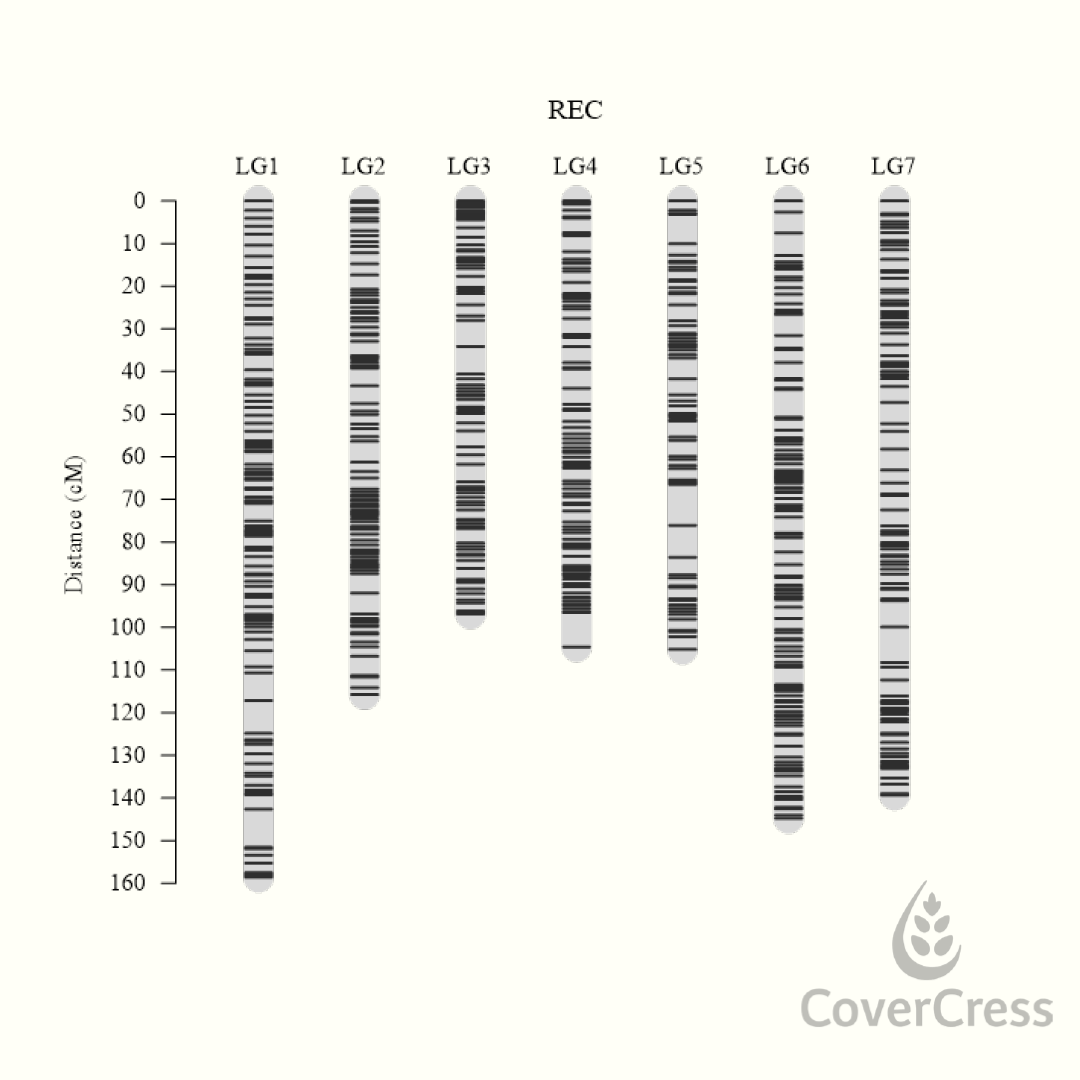
Genotyping around 160 F2 types from their breeding programme using Flex-Seq, they found around 3,500 markers distributed uniformly across the seven chromosomes that segregated in mendelian fashion.
|
|
| Figure 2. Genetic map using Flex-Seq data, Covercress International |
From this they were able to create a genetic map with a total distance between genetic markers of 865.13 cM. This map now informs the company’s germplasm strategies and genomic selection program to avoid creating a genetic bottleneck in a germplasm with narrow diversity. Stacking traits and genes can create linkage blocks that can be detrimental to performance. This map allows CCI to locate and understand recombination frequencies to inform the critical crossing decisions when stacking multiple traits and genes.
Identifying and understanding diversity
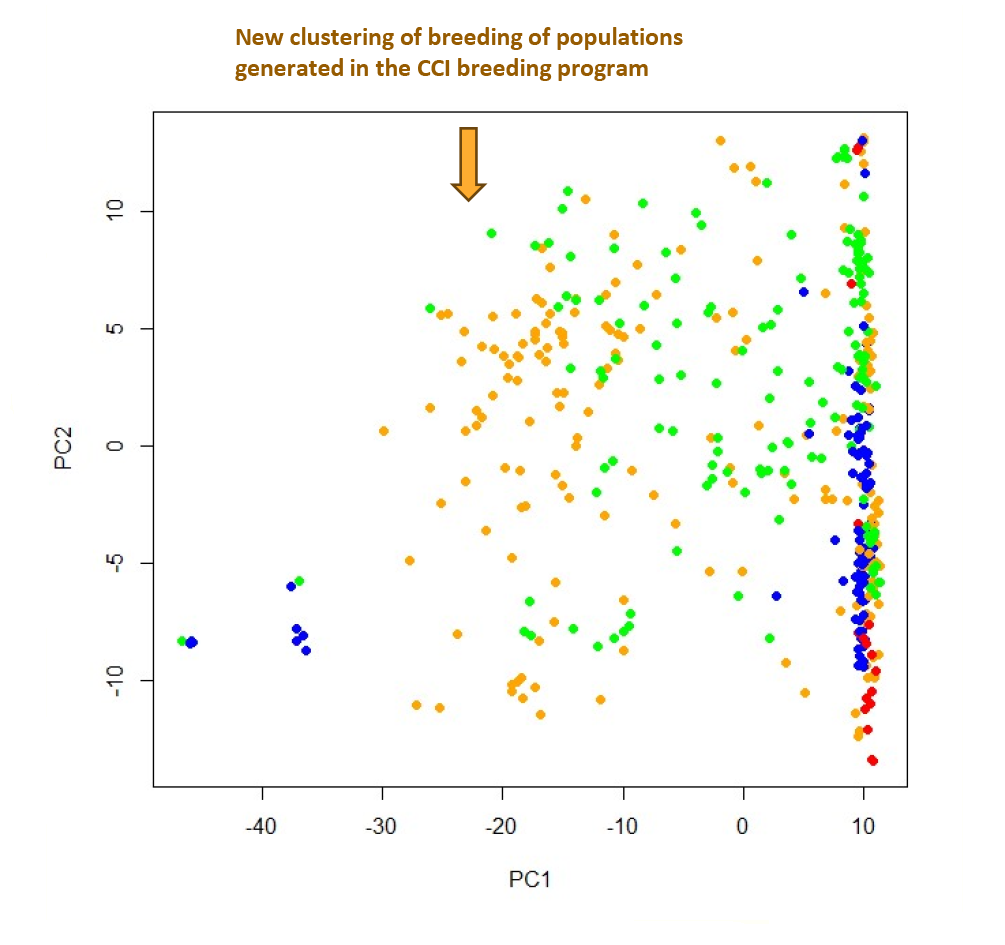
CCI has also been able to use the Flex-Seq platform to understand the diversity of germplasm in the breeding program. When they targeted the 5,000 regions of interest, they were able to segregate 8,000 bi-allelic markers that differentiated 450 samples. The pennycress genetic literature typically shows two major groups. One is a vertical line as shown on the right of the image below and the second is on the extreme left. The one on the right is generally attributed to North American and European cohorts and the one on the left to an Albanian cohort.
|
|
| Figure 3. Diversity markers identified by Flex-Seq data, Covercress International |
The new clustering shows that CCI is creating new germplasm diversity where the varieties are unique to the CoverCress portfolio. More work is in progress to understand the linkage disequilibrium and defining these new populations to leverage for future genomic breeding.
The research team at CCI also wanted to discover if the Flex-Seq data could identify and map quantitative trait loci (QTL) using genome-wide association studies (GWAS). Using the marker data, they were able to do GWAS analysis for the trait and identify associations. They are working on validating these marker trait associations to further refine the QTLs to find genes impacting on the traits evaluated in the program.
“With the Flex-Seq platform we have more data accuracy, reproducibility, better coverage of the genome which will enable genomic selection which will allow us to accelerate the delivery of future products”, said Dr. Chopra.
For more information on the Flex-Seq platform visit our website. Our scientific team are available to discuss your genotyping needs and help advance your genotyping journey, contact us today.