Time to read: 4 minute read
Originally published : Mon, October 2, 2023 @ 3:31 PM
How does genomic selection increase profitability?
Livestock farmers’ and breeders’ profitability is directly related to the quality of their stock. But this can be measured in so many ways: Does the animal put on sufficient weight over the target timeline? How efficiently do they put on that weight? How resistant are they to disease? And so on. Until recently, a farmer had to rely on their farming ‘know-how’ to plough through these complex questions and put their best foot forward. Surely, breeding the largest bull with the largest cow will create supersized offspring; if only it were that simple.
Thankfully today, next generation sequencing (NGS) can help livestock breeders understand their animals at the macromolecule level, taking the roll of the dice out of the process to accelerate vital breeding programmes.
But will the cost eat into profits?
Livestock farmers and breeders operate within tight margins in a highly competitive global market where the end buyers shop around looking for the best price. Over time, those that don’t maximise their livestock’s yield, feeding efficiency and resilience are very likely to lose out to those that do. Investing in genetic gain is a key element to drive profitability and increase sustainability.
A 2021 genome-wide association study in yellow plumage chicken demonstrated how genetics can improve predictions of growth rate traits:1
“The accuracy of genomic best linear unbiased predictions (BLUP) was improved by 22.0 to 70.3% compared to that of the conventional pedigree-based BLUP model. The genomic feature BLUP model further improved the observed prediction accuracy by 13.8 to 15.2% compared to the genomic BLUP model.”
The study concluded that “although each SNP has only a minor effect, the accumulation of the effects of many SNPs throughout the genome has a large impact on prediction accuracy. Although prediction accuracy could be improved by increasing marker density, inclusion of a large number of the sites that do not affect the phenotype could have adverse effects on genomic prediction accuracy, and preselecting SNPs that contribute to phenotypes can improve prediction accuracy and reduce cost.”

In their analysis of genomic selection for cattle in the USA, Gunian et al. concluded that Holstein and Jersey cattle had benefited the most from genomic selection and recommended that coloured breeds in the USA could significantly benefit from genotyping.2 A study conducted by the US Council on Dairy Cattle Breeding found that “the genomic selection program for dairy cattle in the United States has doubled the rate of genetic gain. Since 2010, the average annual increase in net merit has been $85 compared to $40 during the previous 5 years.”3
Although there have been significant improvements in porcine food efficiency (FE), feed costs continue to be a major challenge for the profitable rearing of pigs In their paper on feeding efficiency, the Department of Animal Science at Dalhousie University in Canada (DAL) found that " there is positive phenotypic and genetic correlation between feeding behaviour and FE traits, indicating their possible role on selection of feed-efficient individuals. A deep understanding of feeding behaviour can help breeders to improve feeding strategies, and thus, increase productivity." 4

DAL concluded that, "introducing new traits for selection of FE can significantly improve the accuracy of prediction in pigs. Martinsen et al. (2015) introduced new FE traits like fat efficiency and lean meat efficiency. Their results indicated that these traits help breeders select animals with high genetic potential for efficient deposition of lean meat at low feed costs. The information derived from the integration such as biomarkers and gene networks or causal genes and variants can be incorporated into genomic selection programs to achieve higher accuracy and genetic gain in the pig industry." 4
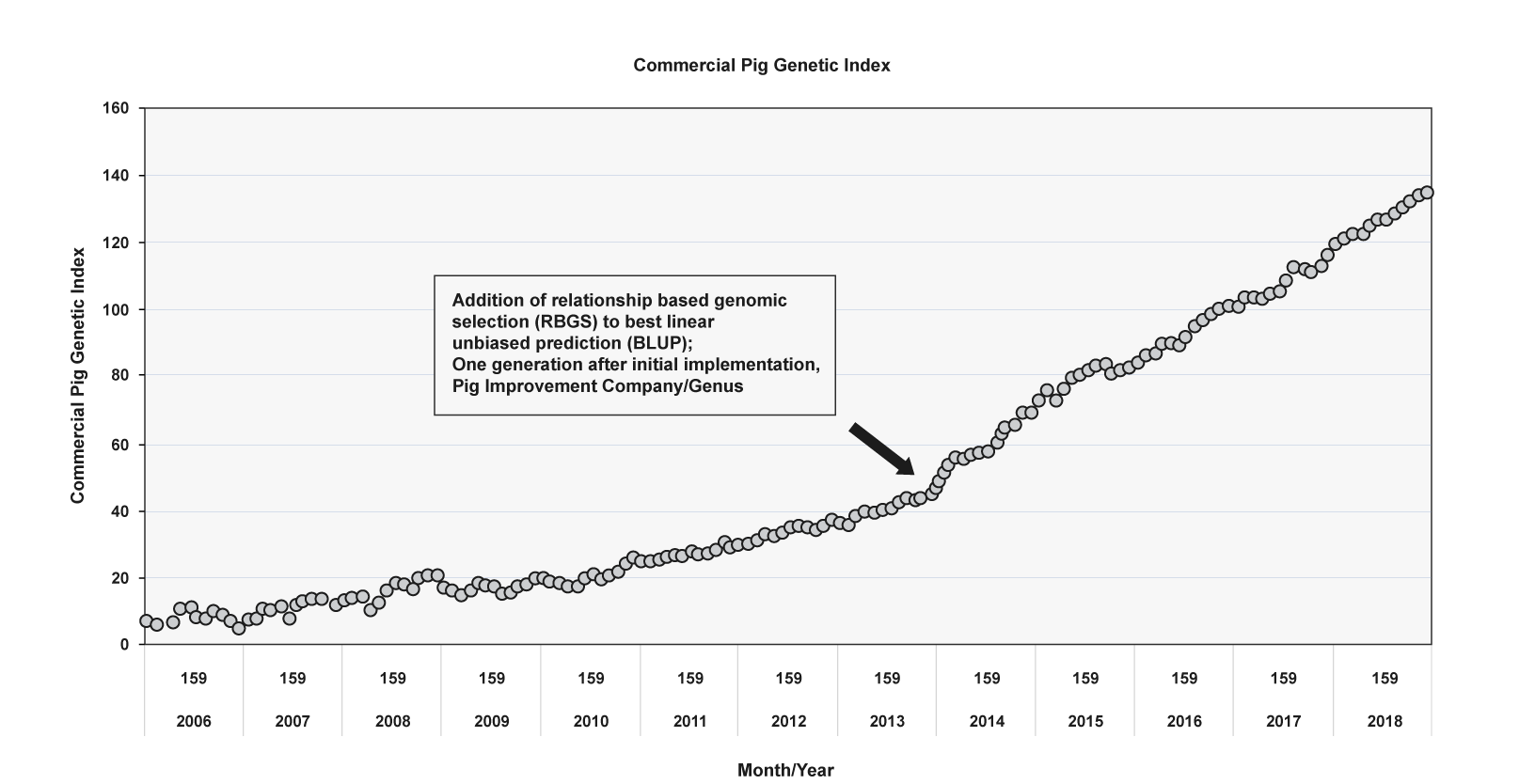
The Pig Improvement Company has mapped its genetic gain index over time, which demonstrates the added value since switching to relationship-based genomic selection in 2013. The graph below shows a doubling of genetic gain when using relationship-based genomic selection versus pedigree selection. PIC have calculated that their Relationship-Based Genomic Selection programme has led to a profit improvement of £ 2.70 to £ 3.10 per pig per year due to a 35% increase in genetic progress per year for all traits.5
It works, but isn’t targeted genotyping via NGS expensive and inflexible?
Flex-Seq™ high-throughput targeted genotyping by sequencing offers a degree of cost flexibility to reach your genotyping and breeding goals, based on the size of the marker panels selected:
- up to 5K SNPs with Flex-Seq LD
- 7K-10K SNPs with Flex-Seq MD
- up to 30K SNPs with Flex-Seq HD
Flex-Seq pre-designed, off-the-shelf panels for porcine, chicken and cattle have flexibility designed in, so changing target markers is a straightforward process. Panels can be adapted easily and quickly to meet genotyping goals to provide a more cost-effective adaptive solution than fixed arrays.
Here's how it works
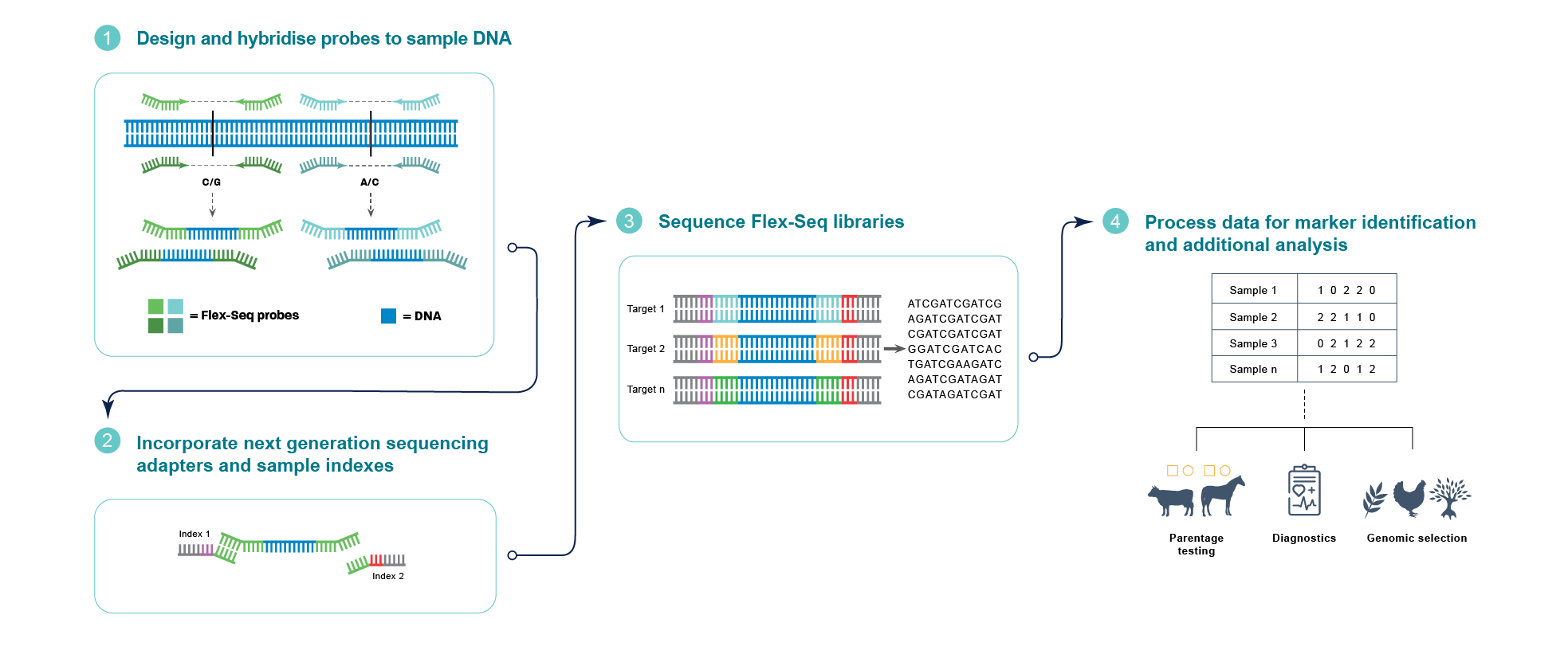 |
| Figure 2 – Illustrative example of the Flex-Seq protocol, highlighting the probe hybridisation to two hypothetical targets and their path via NGS sequencing to genotypic information. |
The probe hybridisation delivers more uniform, highly specific and on-target data than multiplex PCR and allows more diversity in target regions. The use of targeted genotyping by sequence targets the areas of interest, reducing wastage and the data sets are also legacy compatible. The panels can be adjusted over time as scientists travel further in their genotyping journey to look at new areas of interest and can be adjusted for marker density. All of this, combined with an ultra-high-throughput genotyping workflow, creates a cost-effective platform to help achieve genotyping for improved breeding goals.
The cattle, porcine and chicken panels show a high degree of uniformity across the genotyped individuals.
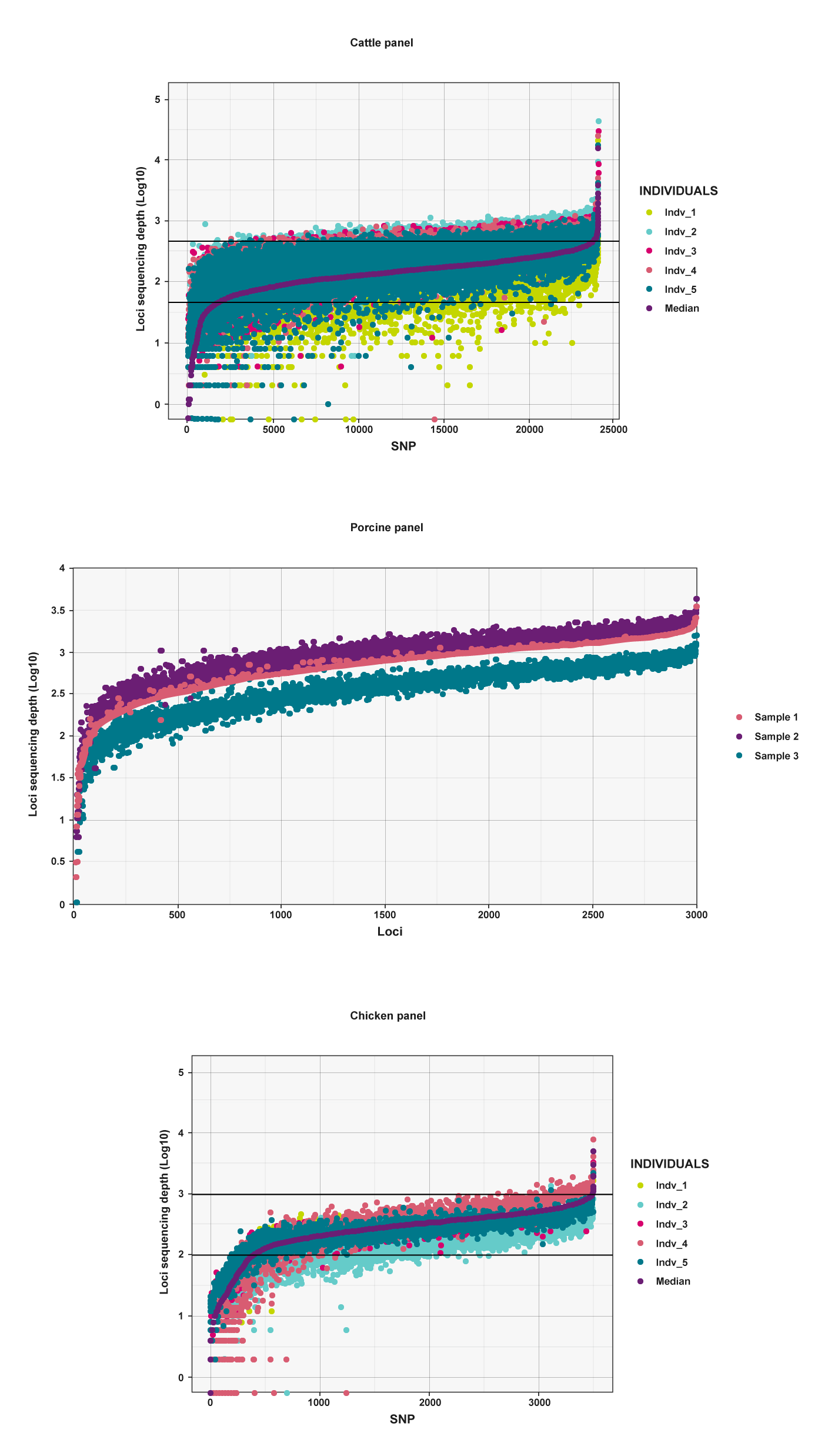
| Figure 3 – Representative loci recovery and uniformity for panels in A) Cattle, B) Porcine, and C) Chicken. The X-axis represents the loci targeted in each panel and the Y-axis represents the amount of sequencing data (log 10 transformed) that each loci received. Data for up to five representative samples are shown for each panel alongside the median. |
View all of our pre-designed industry standard Flex-Seq panels.
For further details on the Flex-Seq panels, watch the webinar with Dr Leandro Neves discussing in detail how Flex-Seq works in animal genotyping.
To request a SNP list for the porcine (Sus scrofa), cattle (Bos taurus) or chicken (Gallus gallus) panels, talk to us today. Our experienced scientists are on hand to work with you to achieve your breeding and genotyping goals. Talk to us today to accelerate your genotyping journey.
Genotyping tools transforming agriculture
References
- Yang, R., et al. Genomewide association study and genomic prediction for growth traits in yellow-plumage chicken using genotyping-by-sequencing. Genet Sel Evol 53, 82 (2021). https://doi.org/10.1186/s12711-021-00672-9
- Guinan, F.L., et al. Changes in genetic trends in US dairy cattle since the implementation of genomic selection. Journal of Dairy Science, Volume 106, Issue 2, 2023,Pages 1110-1129,ISSN 0022-0302, https://doi.org/10.3168/jds.2022-22205
- Wiggans, G. R., et al. Genomic selection in United States dairy cattle. Frontiers in Genetics,Vol 13,2022, https://doi.org/10.3389/fgene.2022.994466
- Pourya, D., et al. Application of Genetic, Genomic and Biological Pathways in Improvement of Swine Feed Efficiency. Frontiers in Genetics, Vol 13, 2022, https://doi.org/10.3389/fgene.2022.903733
- Pig Improvement Company website. Never Stop Improving: Genetic Improvement in the Pig Industry - PIC UK
- Fig 1. Boyd, R.D., et al. Review: innovation through research in the North American pork industry. Science Direct, Vol 13, 2019, https://doi.org/10.1017/S1751731119001915