Originally published : Mon, April 12, 2021 @ 9:16 PM
Updated : Thu, March 9, 2023 @ 5:34 PM
Researchers have identified several variants of SARS-CoV-2 since they sequenced the original viral strain responsible for the COVID-19 pandemic from clinical samples in Wuhan, China. Four variants, UK (B1.1.7), B1.1.7 +E484K, South African (B1.351, 501v2) and Brazilian (B1.1.28, P1), are of particular concern. The United States Center for Disease Control and Prevention (CDC) warns that these variants have the potential to1:
- Cause more severe disease symptoms in patients
- Spread disease more rapidly
- Evade vaccine or naturally-derived immunity
- Evade detection when using specific diagnostic tests
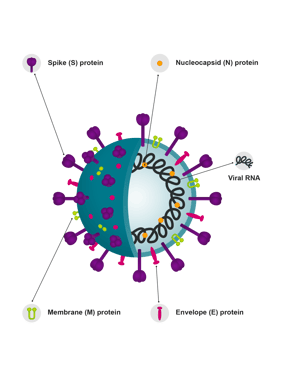
Like other RNA viruses, the SARS-CoV-2 virus mutates - some changes may be inconsequential while others appear advantageous. On average, a typical SARS-CoV-2 virus appears to be acquiring two single-letter mutations per month in its genome.1 Mutations occur along the entire length of the virus's genome, although approximately 50% occur in the spike gene (S-gene). The spike gene encodes the protein associated with entry into host cells and immune response and makes up approximately 13% of the virus's genome.
What are the implications of SARS-CoV-2 variants on diagnostic testing?
SARS-CoV-2 variants are a concern for diagnostic testing because they can evade detection by polymerase chain reaction (PCR) assays currently being widely used to diagnose COVID-19. PCR technology allows amplification of a particular region of a virus's genome and relies on primers specific to each nucleotide sequence targeted. A fluorescent probe, located between the primers, is hydrolysed during the PCR reaction and emits a fluorescent signal that is quantified to indicate the presence or absence of viral RNA.
Because of the probes and primers' specificity to nucleotide sequences, mutations in the genome can create a challenge to obtaining accurate viral detection. The degree to which an assay is affected depends upon the location and type of mutation. For example, research describes the catastrophic failure of one RT-qPCR assay to detect the B1.1.7 variant of the SARS-CoV-2 S-gene.2 In this situation, a deletion of two amino acids from the spike protein in the B1.1.7 variant resulted in the loss of six bases, ultimately helping the virus evade detection using any RT-qPCR assay targeting that region.
What should assay developers, manufacturers and lab testing facilities consider to ensure accurate testing?
The example above illustrates why understanding the biology of SARS-CoV-2 variants is critical for achieving accurate testing results and diagnosis of COVID-19. Stringent sequence screening is ensuring that currently-applied assays do not target genomic regions containing known mutations. Worldwide, many assays target the SARS-CoV-2 nucleocapsid (N-gene), specifically the N1 and N2 areas of the genome. When compared to the S-gene, researchers have identified fewer mutations in the N-gene. Furthermore, few of the mutations identified in the four SARS-CoV-2 variants of concern contain mutations in either of the target regions of the N1 or N2 PCR assays.
However, researchers have identified a mutation within the probe sequence for the N2 assay. The mutation has appeared in 0.22% of virus sequences submitted to the Global initiative on sharing all influenza data (GISAID) to the end of January 2021. Researchers demonstrated that a commercial assay targeting the N-gene failed to detect the mutant viral material. In contrast, the CDC 2019_nCOV_N2 probe detected viral RNA containing the mutation with nearly no sensitivity loss.3 This example illustrates that the characteristics and position of arising mutations can impact detection differently, depending on the assay.
How is LGC, Biosearch Technologies™ ensuring assay integrity and accurate SARS-CoV-2 viral detection?
 Biosearch Technologies is closely monitoring SARS-CoV-2 variants and developing proactive strategies to continue to provide mission-critical tools and resources to our customers. To stay one step ahead of evolving SARS-CoV-2 viruses, we are:
Biosearch Technologies is closely monitoring SARS-CoV-2 variants and developing proactive strategies to continue to provide mission-critical tools and resources to our customers. To stay one step ahead of evolving SARS-CoV-2 viruses, we are:
- Conducting fortnightly screens of the N1 and N2 assays against the sequences held in GISAID to alert us of any prevalent mutations that could adversely impact an assay's performance.
- Creating protocols that will facilitate rigorous assay functionality testing if a variant of concern develops.
- Assessing alternative diagnostic assay combinations and proactively preparing them for deployment, if the need arises.
- Developing a range of SARS-CoV-2 Variant ValuPanel™ assays of probes and primers for the qualitative detection of emerging virus mutations.
- Launching NxSeq SARS-CoV-2 Whole Genome Library Kit, a fast and efficient, amplicon-based SARS-CoV-2 whole genome library preparation that’s compatible across Illumina sequencers.
- Offering a whole-genome sequencing service* to identify emerging virus variants.
In addition to these actions, our teams will continue to remain engaged in discussions and share relevant research regarding SARS-CoV-2 variants of concern. As the landscape for testing continues to change, we will ensure that our entire team has the latest information so that our customers can quickly and effectively adapt their strategies as needed.
Even though more people are receiving the COVID-19 vaccine, we cannot become complacent in our efforts to identify and manage new variants that can threaten public health. Genotyping new strains of the virus to develop the most accurate testing assays and using RT-qPCR analysis to quickly and accurately diagnose cases are crucial steps for managing evolving SARS-CoV-2 variants. To proactively identify new and novel variants worldwide, widespread and consistent adoption of SARS-CoV-2 genome sequencing protocols is essential. As more labs sequence a percentage of positive patient samples, we can better understand and track how the virus is mutating over time and across geographies. That critical information will help guide future testing and treatment plans as the virus evolves.
*Serving EMEA region only.
References
- Callaway, E. Making Sense of Coronavirus Mutations. Nature. 585(7824):174-177. https://media.nature.com/original/magazine-assets/d41586-020-02544-6/d41586-020-02544-6.pdf. Published 2020. Accessed April,1 2021.
- Centers for Disease Control and Prevention. Genomic Surveillance for SARS-CoV-2 Variants. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance.html. Published 2021. Accessed March 18, 2021.
- Public Health England. Investigation of Novel SARS-CoV-2 Variant, Variant of Concern 202012/01. Technical Briefing 4. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959359/Variant_of_Concern_VOC_202012_01_Technical_Briefing_4.pdf. Published January 2021. Accessed March 18, 2021.
- Ziegler K, Steininger P, Ziegler R et al. SARS-CoV-2 Samples May Escape Detection Because of a Single Point Mutation in the N Gene. Euro Surveill. 25(39):pii=2001650.
DOI: 10.2807/1560-7917.ES.2020.25.39.2001650. Published 2020. Accessed March 18, 2021.


