Originally published : Thu, May 14, 2015 @ 3:39 PM
Updated : Fri, November 13, 2015 @ 10:27 PM
This past April, Biosearch attended the American Association of Cancer Research (AACR) conference in Philadelphia, PA where, in collaboration with Byung Chul Kim from Clinomics, Inc, we shed light on the detection of fusion genes by means of Stellaris® RNA FISH. Gene fusions have the potential to activate otherwise silent signaling enzymes, such as the anaplastic lymphoma receptor tyrosine kinase (ALK), leading to significant numbers of lung and other related cancers. This particular gene fusion occurs from inversions on chromosome 2 (inv2(p21p23)), fusing the EML4 and ALK genes and connecting the promoter and 5' half of EML4 (active in normal lung tissue) together with the 3' and kinase encoding half of the otherwise silent ALK gene.
Stellaris FISH is a remarkable tool for illustrating precisely what is happening at the transcriptional level. An identical technique, iceFISH™, targets intron chromosomal expression and serves to measure gene expression and chromosome structure simultaneously on single chromosomes. Since most post-transcriptional processing, including pre-mRNA splicing, occurs co-transcriptionally, introns can be used as a location proxy for the encoding gene. We utilized these two approaches to detect the EML4-ALK fusion, both of which targeted the different 5' and 3' segments of the pre-mRNAs and mature mRNAs of wild type EML4 and ALK, and EML4-ALK fusions.
The following RNA FISH probe sets targeting pre-mRNAs and mature RNAs were designed accordingly–
- EML4 5’ intronic Quasar 570
- EML4 3’ intronic Quasar 670
- ALK 5’ intronic Cal Fluor Red 610
- ALK 3’ intronic Fluorescein
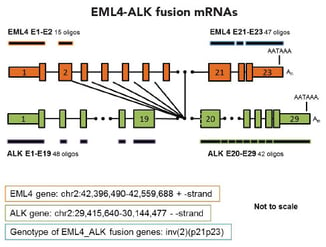
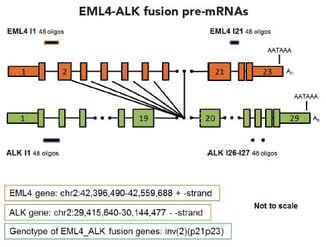
Fig. 1 - Schematic of Stellaris RNA FISH probe sets for EML4-ALK fusion pre-mRNA and mRNAs.To view larger images of the schematics, please refer to Figure 2 in the fusion detection poster.
Each set of probes was labeled with spectrally distinct fluorophores, making it possible to detect multiple RNA targets simultaneously. We used the non-small cell lung cancer (NSCLC) adenocarcinoma H2228 cell line, which carries one wild type and two mutant alleles on chromosome 2. The mutant alleles contain two different inversions, both presumed to be active in the cell for the testing of the probes sets to detect wild type and fusion pre-mRNAs and mature mRNAs.
Through iceFISH in H2228 cells, we found EML4 5’ intronic RNA co-localized with EML4 3’ intronic RNA, thus verifying the presence of the wild type EML4 gene. Additionally, the presence of the EML4-ALK fusion gene was also confirmed but by the co-localization of EML4 5’ intronic RNA with ALK 3’ intronic RNA (Fig. 2). Next, we targeted the mature RNAs and found co-localized signals for wild type cytoplasmic EML4-mRNAs and EML4-ALK fusion mRNAs (Fig. 3).
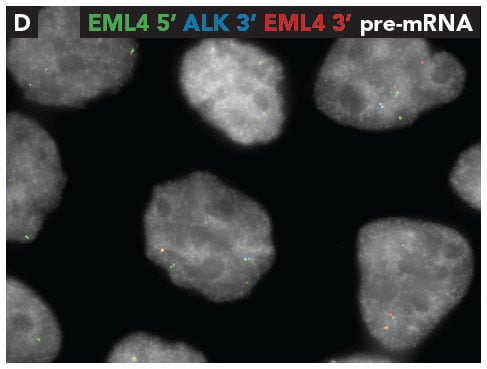
Fig. 2 - Co-localization of intronic EML4 5' RNA (green) with the intronic EML4 3' RNA (red) and EML4 5' RNA (green) with ALK 3' RNA (blue). To view related images please refer to Figure 4 in the fusion detection poster.
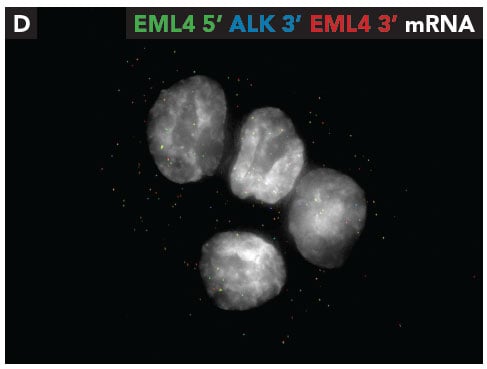
Fig. 3 - Co-localization of EML4 5' mRNA (green) with EML4 3' mRNA (red) and EML4 5' mRNA (green) with ALK 3' mRNA (blue). To view related images please refer to Figure 3 in the fusion detection poster.
We also sought to detect the EML4-ALK fusion in circulating tumor cells (CTCs). CTCs are cells that have sloughed off from cancerous tissue and have moved into the vasculature of the bloodstream. Circulating through the bloodstream, they travel alone, in pairs, or in clusters. Regardless of where they may end up, they share one key aspect--their origin to the tumor cell1,2.
The maturation of CTC isolation technology over the past decade has lead to an accelerated understanding of the correlation between the number of CTCs found in blood and a patient’s disease progression3. Isolated CTCs can further undergo genetic characterization through single-cell analysis techniques, such as fluorescence in situ hybridization, DNA, or RNA assays. Characterization of CTC’s can elucidate prognostic and predictive factors. The exploration of biological processes, such as dissemination, drug resistance or therapy-induced cell death, can influence a patient’s therapy, potentially improving their prognosis and quality of life4. Ultimately, characterization at the molecular level is prudent for understanding what aspects of a tumor can be inferred from CTCs. When applying Stellaris RNA FISH to CTCs, we were once again able to distinguish cells that tested positive for the wild type EML4 gene alongside the EML4-ALK fusion gene.
Stellaris RNA FISH is a complementary method to traditional expression assay techniques. Stellaris FISH exposes the precise location of single molecules of RNA such that they can be quantified. iceFISH provides not only a view of where the gene expression is occurring within the chromosome, but also pinpoints transcriptionally active genes in particular cells at a particular time. Assays for EML4 and ALK iceFISH are available on our website. We applied both methodologies (targeting mature and immature RNA) to demonstrate the effectiveness of RNA FISH in detecting the presence of an EML4-ALK fusion gene. We anticipate further discovery and detection of fusion genes as their implication and role in cancers continues to be defined.
References
- Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Australian Medical Journal. 1869
- Miller C, Doyle G, Terstappen L. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. Journal of Oncology. 2010
- Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. New England Journal of Medicine. 2004
- S. Sleijfer, et al. Circulating tumour cell detection on its way to routine diagnostic implementation? European Journal of Cancer. 2008