Originally published : Wed, June 24, 2020 @ 4:23 PM

Updated : Tue, July 14, 2020 @ 1:30 PM
Brassica rapa crops are important sources of food, feed, and fuel in many parts of the world and provide a unique opportunity for investigating plant genome evolution. In 2018, the Food and Agricultural Organization reported a global production of cabbages and other brassicas to be more than 103 million tonnes.1 Disease and environmental stress can reduce crop yield and quality, so plant breeders are using genome-wide molecular markers like SNPs to make a background selection and identify target genes involved in high nutritional quality, disease resistance and yield in order to speed up the breeding process in Brassica rapa crops.
SNPs also have great value in the seed quality testing industry as there is an urgent need for an accurate, simple and fast method to test the purity and authenticity of seeds and to protect intellectual property rights of the breeders.
The research group led by Dr. Shuancang Yu from the Beijing Vegetable Research Center (BVRC) successfully identified a set of 568 SNPs in Brassica rapa crops using KASP™ genotyping technology, which they found to be an economic and accurate approach for germplasm analysis and breeding application. In their study of genetic diversity, they discovered that KASP was “the most adaptable, efficient and cost-effective genotyping system.”
 Research group led by Dr. Shuancang Yu from the Beijing Vegetable Research Center (BVRC). From left to right: Shuancang Yu (Deputy Director, BVRC), Tongbing Su (Associate Professor, BVRC), Peirong Li (Assistant Professor, BVRC).
Research group led by Dr. Shuancang Yu from the Beijing Vegetable Research Center (BVRC). From left to right: Shuancang Yu (Deputy Director, BVRC), Tongbing Su (Associate Professor, BVRC), Peirong Li (Assistant Professor, BVRC).


 Researchers from BVRC (from left to right - Pan Li, Assistant Professor; Xiangjing Li, researcher; Tongbing Su, Associate Professor) demonstrating the use of LGC, Biosearch Technologies’ K-pette, Fusion™ and Hydrocycler™ to replicate samples, seal and thermal cycle multiple 1536 plates in a water bath as part of the high-throughput end-point SNP genotyping workflow using KASP technology.
Researchers from BVRC (from left to right - Pan Li, Assistant Professor; Xiangjing Li, researcher; Tongbing Su, Associate Professor) demonstrating the use of LGC, Biosearch Technologies’ K-pette, Fusion™ and Hydrocycler™ to replicate samples, seal and thermal cycle multiple 1536 plates in a water bath as part of the high-throughput end-point SNP genotyping workflow using KASP technology.
What is KASP?
PCR-based KASP (Kompetitive Allele-Specific PCR) genotyping technology is a homogeneous, fluorescence (FRET) based assay that enables accurate bi-allelic discrimination of known SNPs and InDels. Biosearch Technologies is the only provider of the original patented KASP technology, while providing end-to-end solutions and best-in-class technical consultation to support the entirety of the genotyping workflow. Our automated, high-throughput sample extraction, large-scale liquid dispensing and PCR instrumentation, and data analysis software are all optimised around our KASP genotyping chemistry, and is the same system we used for our service labs.
KASP is accurate, reproducible, scalable and economical
With the KASP genotyping system, the BVRC research group experienced:
- Superb accuracy and dependable performance - accuracy >99.8%* and industry-leading SNP and InDel assay conversion rate (>90%).
- Easy-to-use technology that was scalable - Flexible primer design combined with Biosearch Technologies' 15+ years of expertise produced a successful assay design. KASP chemistry optimised on our high-throughput genotyping automation platform, SNPline™, enabled BVRC to easily scale up to 2-3 million data points per year with this streamlined workflow.
- Affordability - BVRC commented in their publication that “many technologies exist for use in SNP genotyping analyses...with KASP being the most highly adaptable, efficient and cost‐effective system”.
Noteworthy data from the BVRC study
Below are example data sets to demonstrate how this core set of SNPs were identified with the robust KASP genotyping technology.
In this study, 3,183 SNPs were selected for KASPar assays development, and 568 were finally converted and selected for Brassica rapa germplasm characterisation.
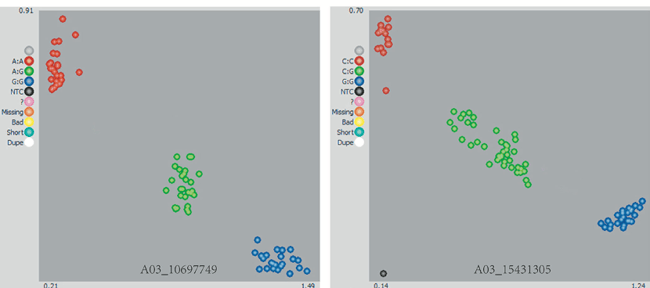 Figure 1. Development of SNP markers from Brassica rapa inbred lines for KASP genotyping. Single‐nucleotide polymorphisms were automatically called for AA, AB and BB genotypes. Red dots are homozygous for one allele, blue dots are homozygous for the second allele and green dots are the heterozygous allele.
Figure 1. Development of SNP markers from Brassica rapa inbred lines for KASP genotyping. Single‐nucleotide polymorphisms were automatically called for AA, AB and BB genotypes. Red dots are homozygous for one allele, blue dots are homozygous for the second allele and green dots are the heterozygous allele.
Figure 2. Distribution of 568 SNP marker loci on the 10 chromosomes of Brassica rapa.
Based on the SNP dataset, structure and principal coordinates analysis, and neighbor-joining phylogenetic methods were used to examine the population structure and gave highly consistent results. The 231 accessions were divided into the four primary subspecies, representing 99 accessions from B. rapa ssp. pekinensis, 85 from B. rapa ssp. chinensis, 30 from B. rapa ssp. rapifera, and 17 from B. rapa ssp. oleifera and were further subdivided into 12 lower-order clusters according to different morphotypes (see below).
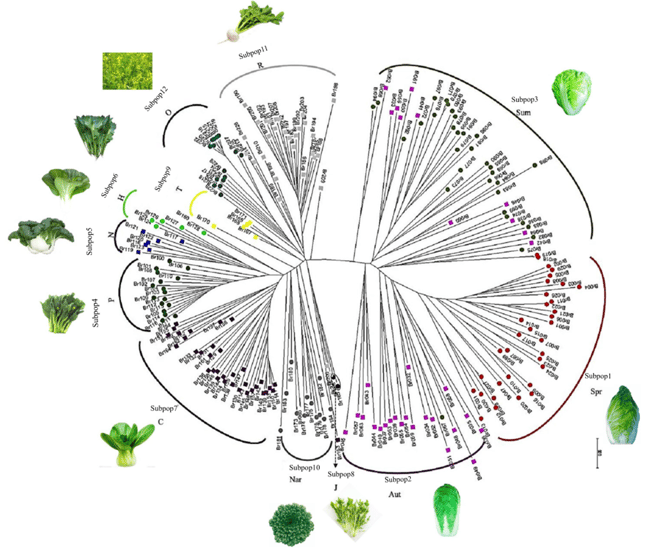 Figure 3. Analyses of population structure in B. rapa. Populations structure analysis. All 231 germplasm accessions were further divided into 12 subpopulations. Pop1, ssp. pekinensis; Pop2, ssp. chinensis; Pop3, ssp. rapifera; Pop4, ssp. oleifera. Subpop1, spring-type Chinese cabbage lines of ssp. pekinensis(Spr);Subpop2,autumn-type(Aut);Subpop3,summer-type (Sum); Subpop4, ssp. chinensis var. parachinensis (P); Subpop5, ssp. chinensis var. communis (naibaicai) (N); Subpop6, ssp. chinensis var. communis (heiyebaicai) (H); Subpop7, ssp. chinensis var. communis (C); Subpop8, ssp. chinensis var. japonica (J); Subpop9, ssp. chinensis var. taicai (T); Subpop10,Nar,ssp.chinensis var. narinosa(Nar); Subpop11, ssp. rapifera (R); Subpop12, ssp. oleifera (O)
Figure 3. Analyses of population structure in B. rapa. Populations structure analysis. All 231 germplasm accessions were further divided into 12 subpopulations. Pop1, ssp. pekinensis; Pop2, ssp. chinensis; Pop3, ssp. rapifera; Pop4, ssp. oleifera. Subpop1, spring-type Chinese cabbage lines of ssp. pekinensis(Spr);Subpop2,autumn-type(Aut);Subpop3,summer-type (Sum); Subpop4, ssp. chinensis var. parachinensis (P); Subpop5, ssp. chinensis var. communis (naibaicai) (N); Subpop6, ssp. chinensis var. communis (heiyebaicai) (H); Subpop7, ssp. chinensis var. communis (C); Subpop8, ssp. chinensis var. japonica (J); Subpop9, ssp. chinensis var. taicai (T); Subpop10,Nar,ssp.chinensis var. narinosa(Nar); Subpop11, ssp. rapifera (R); Subpop12, ssp. oleifera (O)
.
Based on the above, the researchers selected 50 and 60 core SNPs that can provide sufficient genetic polymorphism information of Brassica rapa subsp. chinensis and Brassica rapa L. subsp. pekinensis, respectively, for further seed purity and authenticity testing, germplasm characterisation and molecular marker‐assisted breeding.
Interested in testing KASP genotyping chemistry in your lab?

About Beijing Vegetable Research Center (BVRC)
Beijing Vegetable Research Center(BVRC), Beijing Academy of Agriculture and Forestry Sciences was founded in 1958, which is a scientific research institution affiliated to Beijing municipal government, It was officially entitled as the National Engineering Research Center for Vegetables by The Ministry of Science and Technology of People’s Republic of China in 1995. The Center is located in Beijing with a nationwide perspective, the research targets on the forefront of vegetable science and technology, and providing comprehensive scientific and technical support to ensure efficient and sustainable development of vegetable industry in China. The center was the first to introduce KASP™ genotyping technology in China, and applied it widely in vegetable crops breeding. In the past few years, more than 500 vegetable varieties have been released by BVRC, which not only planted across China, but also exported to 15 countries. BVRC has become one of the most distinguished institutions, and outstanding professional vegetable research institution in China. The Center has developed bilateral and crosswise international cooperation of science and technology with Japan, USA, United Kingdom, Germany, France, and more than other ten countries. Moreover, sponsored by Chinese government, the Center has continually conducted the international training courses of vegetable science to more than 40 developing countries in recent decades, accumulatively trained more than 800 foreign managerial technicalist of vegetable science and technology,and gains a great reputation in the world.
References:
Su, T., Li, P., Yang, J. et al. Development of cost-effective single nucleotide polymorphism marker assays for genetic diversity analysis in Brassica rapa. Mol Breeding 38, 42 (2018). https://doi.org/10.1007/s11032-018-0795-0
Li, P. et al. 2019. Development of a core set of KASP markers for assaying genetic diversity in Brassica rapa subsp. chinensis Makino. Plant Breeding. 138(3):309-324.doi.org/10.1111/pbr.12686
Li, P., Su, T., Yu, S. et al. Identification and development of a core set of informative genic SNP markers for assaying genetic diversity in Chinese cabbage. Hortic. Environ. Biotechnol. 60, 411–425 (2019). https://doi.org/10.1007/s13580-019-00138-4
Food and Agriculture Organization Crop Database.http://www.fao.org/faostat/en/#data/QC


