Originally published : Wed, January 13, 2016 @ 11:06 PM
Updated : Tue, September 27, 2022 @ 2:32 PM
Stellaris® RNA FISH is a novel technology that allows for the detection of single or clustered RNAs in fixed cells and tissue. Multiple unique probes with similar stability are designed to hybridize along the length of a single RNA target and upon hybridization, generate a fluorescent signal. LGC Biosearch Technologies offers customers a web-based, free, and easy to use software for Stellaris RNA FISH Probe design. The probe designer takes an input sequence, such as an mRNA coding sequence, and outputs a set of up to 48 probes designed for optimal binding properties.
Typically, a minimum of 25 probes are required for reliable detection. For most long unique RNA targets it is not difficult to reach this number of oligos. However, some particularly short or repetitive targets are more challenging, and it might not be possible to reach the minimum of 25 probes on the first pass. Fortunately, in these cases, there is a series of changes that can be made to the designer settings which will increase the likelihood of generating a full probe set. We hope that the following suggestions are helpful to many of our users who are having difficulty obtaining enough probes for their custom design. While not all targets are compatible with the Stellaris RNA FISH technology, a challenging target may be successfully visualized with some of the following design strategies.
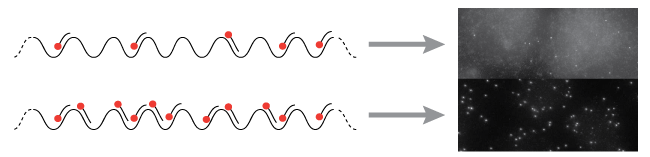
Targets with fewer probes binding will have dimmer signal compared to the surrounding area
Design Issue: Not Enough Probes
How to tell if you don’t have enough probes
Once you’ve submitted your target sequence with your first round of designer settings, you will be taken to the designer results page. On this page, you should see the phrase “Count [#] (Maximum 48 probes per order.)” If that number is below 25, you are below our recommended number of probes for a probe set. Additionally, the following warning message should appear “ATTENTION: We recommend a minimum of 25 oligos for a single probe set.”
Possible reasons why your design doesn’t have enough probes
Placement of the oligo-probes is firstly based on thermodynamic parameters to ensure similar binding characteristics. Sequences with a skewed GC-content (high or low) will not be targeted with a probe. Secondly, repetitive sequences are masked in the input sequence and the stringency can be adjusted (levels 1-5).
Whether your low probe number is caused by incompatible GC-content or repeats, follow the steps below to try and solve this issue. We will be reviewing a number steps to address both of these root causes, as most low count probe sets benefit from both kinds of adjustments. Each step below should increase or maintain the number of probes depending on the specific issue you are having. Follow the steps sequentially until you reach a sufficient number of probes.
Design Troubleshooting
How to improve your probe GC-content and what can be done if you don’t initially have enough probes?
Here, we will be trying to increase probe set count by solving the skewed GC-content and some Sequence Masking issues by changing the designer settings on the main design page. We will address both increasing sequence eligibility and lowering masking settings. Sequence Eligibility will not alter the sensitivity and specificity of your Stellaris Custom Probe Set, so this should be done before working on masking which may affect the specificity and sensitivity of your probe set.
Before you start troubleshooting your design by changing designer settings, double check that the sequence you have entered is indeed the desired target sequence. This may seem like a no-brainer, but omitting parts of your target or having the wrong sequence information can quickly turn an easy design experience into a frustrating one. So take the time to carefully select your input sequence.
Increasing the input sequence length is a fast and easy way to increase the number of probes in your design. While you’re reviewing your input sequence, look to see if your submitted sequence can be expanded by including other sequence information such as the 5’ or 3’ UTR for mRNA targets or additional introns for intron FISH (iceFISH). For iceFISH, you can also include a small stretch (up to 10 nt) of the surrounding exons to give the designer a little more room to fit a few more probes. Keep in mind that probes designed to the junction between introns and exons will only bind the pre-mRNA, and not the mature mRNA.
Run your sequence through the designer again to see if your expanded sequence gives you enough probes. If not, continue the troubleshooting steps.
Consideration #1: Dealing with Skewed Sequence GC-ratio
Some parts of your sequence simply aren’t eligible to design probes against based on sequence composition. The probe designer selects specific regions for probe binding using a predicted melting temperature range. This ensures all probes perform similarly during hybridization. Since melting temperature is partially dependent on GC content, targets with poorly distributed GC content will be difficult for probe design.
Step 1: Reduce Probe Spacing
The first thing that can be done, is to adjust probe spacing. Probe spacing refers to the minimum number of nucleotides between probe binding sites along your target. The default setting is 2, but can be reduced to 1 without affecting probe binding efficiency. This is both a quick and easy way to immediately increase the probe number, but is also an important preparation step for steps 2 and 3 below. Please note that decreasing probe spacing should NOT BE DONE if you are planning on using the CAL Fluor Red 635 dye. This particular dye requires a two nucleotide spacing because it attached with a different linker to the oligo. Changing this setting should have no effect on the final performance of your probe set for the other Biosearch dyes.
Step 2: Vary Probe Length
Second, varying the probe length between 18 and 22 nucleotides should be tried. High GC-content sequences, such as many 5’ UTRs, are better targeted by 18- and 19-mers, whereas AT-rich sequences, such as 3’ UTRs are better targeted by 21- and 22-mers. The best setting for your specific sequence will be dependent on the GC content. Try each length starting with 18 and moving up to 22 to see which, if any, of these length choices increase your probe set sufficiently.
Step 3: Mixed-mer Probe Set
If your target has non-uniform GC content with distinct GC-rich and AT-rich regions, you can also create a mixed length probe set. It is possible to create a larger set of mixed-mer probes by combining NON-OVERLAPPING probes from different design sets of different probe lengths.
Simply iterate your designs with varying probe lengths. Copy down the probe sequences recommended from each of these different length settings, and combine the non-overlapping probes into one set. Your final probe set will now be comprised of probes which will range in length from 18 to 22 nt instead of all being a uniform length.
To view the location of each probe within the design, go to the “Review Design Results” page of the designer and click on “View Input Sequence with Probe Locations.” This should display the locations of the probes and help you combine different designs. Just make sure the probes that you are selecting do not overlap or they will compete with each other.
Currently, mix-mer probe sets must be ordered by submitting the oligos entered into the Stellaris excel order form that can be found on the Stellaris Custom Probe Sets page.

Two designs of different probe lengths combine to create a larger mixed-mer set. More than two designs can be combined to create a mixed-mer set.
Consideration #2: Dealing with too much Masking
The designer requires unique sequence information and will automatically mask simple repeats, species specific repeats, and sequences that appear many times throughout the selected genome. This is done to prevent probes from binding to repetitive or non-unique target sequences and higher background.
Step 4: Reduce the Masking Level to 4, then 3
If the designer is masking too much sequence, reducing the masking level from 5 to 4, and then 3, will relax the designer and make more input sequence available for the probe design. Be sure to check the probe number as you decrease the masking level to ensure that you do not unnecessarily lower the masking level too low while still generating enough probes for your Stellaris RNA FISH probe set. Keep in mind that decreasing the masking level may cause you to lose some protection from targeting pseudogenes and other similar sequences from the genome. Therefore, especially if you are decreasing the masking levels, it is good practice to screen your probe for specificity to the selected transcriptome.
To screen your probe set post-design, affix fasta headers to each oligo sequence (E.g. >Probe_1, etc.) and BLAST them against the transcriptome of the chosen organism. Directions on how to use NCBI’s BLAST tool can be found at the BLAST help page. Probes with 16 or more (assuming 20 nt probes) nucleotides complementarity to non-target RNAs are at risk of hybridizing and should be de-selected from the set. Be sure to note sequence direction. The binding of a few probes to other targets is expected, and does not interfere with the overall performance of the probe set. Stellaris probe imaging relies on multiple probes localizing to a single target to generate a detectable signal. If an off-target gene matches to five or more probes with 16 or more nucleotides, you will need to remove those probes.
Step 5: Reduce the Masking level to 2, then to 1
If the above steps still do not generate a probe set containing 25 or more probes, try reducing the masking level below 3. This masking level removes any species-specific masking. This isn’t necessarily a bad thing as you still can BLAST the individual probes against your relevant transcriptome and remove unwanted probes. It is absolutely critical to BLAST your probe sequences if you have lowered the masking level to 2 or 1, so, especially in this case, this BLAST step should not be seen as optional. See the directions above for BLASTing instructions.
Final Words
If you are still having difficulties with your design, feel free to contact our technical support team with any questions you may have. You can contact us via email at techsupport@biosearchtech.com or over the phone at 1(415)883-8400.

