Image credit: HIV Reverse Transcriptase. RCSB Protein Data Bank. The Wellcome Collection.
Reverse transcriptases (RTs) are enzymes that can generate complementary DNA (cDNA) from an RNA template. RTs are RNA-dependent DNA polymerases, which were initially discovered in retroviruses. They now have widespread applications in various molecular biology techniques used for biomedical research and molecular diagnostics.
How RTs work: structure and function
Retroviral reverse transcriptases have different structures, but share two key elements: a DNA polymerase domain and a Ribonuclease H domain (RNase H).1,2
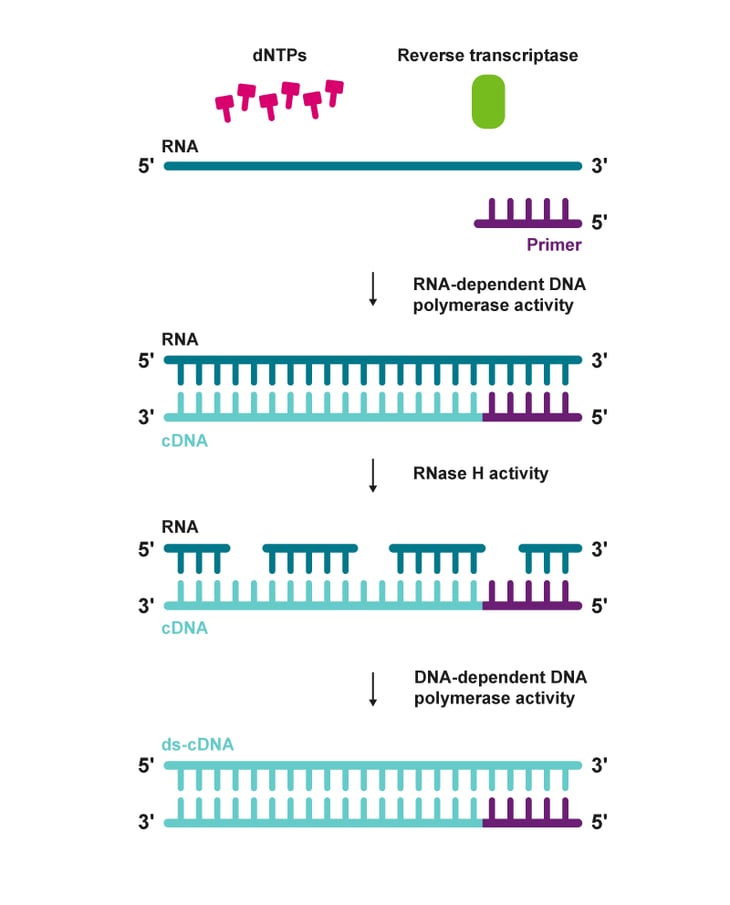
|
| Figure 1: Steps involved in a reverse transcription process |
The DNA polymerase domain can use either RNA or DNA as a template. The RNase H domain cleaves the RNA strand of an RNA:cDNA hybrid to yield a single-stranded cDNA. By combining these two enzymatic functions, reverse transcriptases can synthesise double-stranded DNA from RNA and a primer. (figure 1).2
How to select the most suitable RT for your application?
Most commercially available reverse transcriptases are derived from the avian ameloblastoma virus (AMV) or the Moloney murine leukemia virus (MMLV) and the properties are engineered to suit the application technique.
Reverse transcriptases from different sources vary in the capacity, length and yield of cDNA synthesis. Apart from the length and secondary structure of the target RNA, the key features of RTs to consider when selecting for a molecular biology application are:
- DNA Polymerase activity – Depending on the source organism, the RNA- and DNA-dependent polymerase activity can vary. DNA-dependent polymerase activity, if present, will yield a double-stranded cDNA.
- RNase H activity – RNase H activity is often curtailed to avoid cleavage of the RNA template during polymerisation and improve efficiency by minimising competition with the DNA polymerase active site.
- Thermostability – RNAs with complex secondary structures require higher temperatures for denaturation. Therefore, thermostable reverse transcriptases can allow full-length cDNA synthesis.3
- Processivity – High processivity means the incorporation of more nucleotides in a single binding event and hence a shorter reaction time.
- Fidelity – High fidelity refers to a lower error rate during reverse transcription.
- Terminal nucleotidyl transferase (TdT) activity – When reverse transcriptases reach the 5′ end of the RNA template, they can add 1-3 unspecific nucleotides to the double-stranded DNA:RNA or DNA:DNA substrate.
Considerations for reverse transcriptases in genomics and transcriptomics
Reverse transcriptases are a valuable tool for genomics and gene expression studies because they allow for the conversion of intracellular RNA to more stable cDNA, which can then be studied or cloned for further investigation. RTs are commonly used in a range of molecular biology workflows, including polymerase chain reaction (PCR), cloning and sequencing. Here we discuss the most common techniques that utilise reverse transcriptase enzymes and the desirable properties of each technique.
cDNA libraries
One of the first applications of reverse transcriptases was to construct cDNA libraries where the total mRNA transcripts (expressed genes) in eukaryotic cells were converted to cDNA and inserted into suitable vectors. cDNA libraries provide insights into gene functionality in various physiological conditions and are useful for expressing eukaryotic proteins in prokaryotic cells. RNase H enzyme is used in the workflow to nick the RNA template to yield double-stranded cDNAs.4 Moreover, a thermostable reverse transcriptase with muted RNase H activity and high processivity (like the EpiScript RNase H- Reverse Transcriptase Kit) is preferred for library preparations to capture low abundance mRNAs, full-length mRNAs and RNAs with complex secondary structures.
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
RT-PCR is one of the most routinely used techniques in genomic research as well as molecular diagnostics. RT-PCR involves the conversion of the target mRNA population in a cell to cDNA, followed by the amplification of cDNA (figure 2). RT-PCR is used to detect or amplify low-abundance RNA and requires thermostable reverse transcriptases with high efficiency. Modification of RT-PCR such as quantitative RT-PCR, Rapid amplification of cDNA ends (RACE) and Reverse transcription loop-mediated isothermal amplification (RT-LAMP) also work on a similar principle.
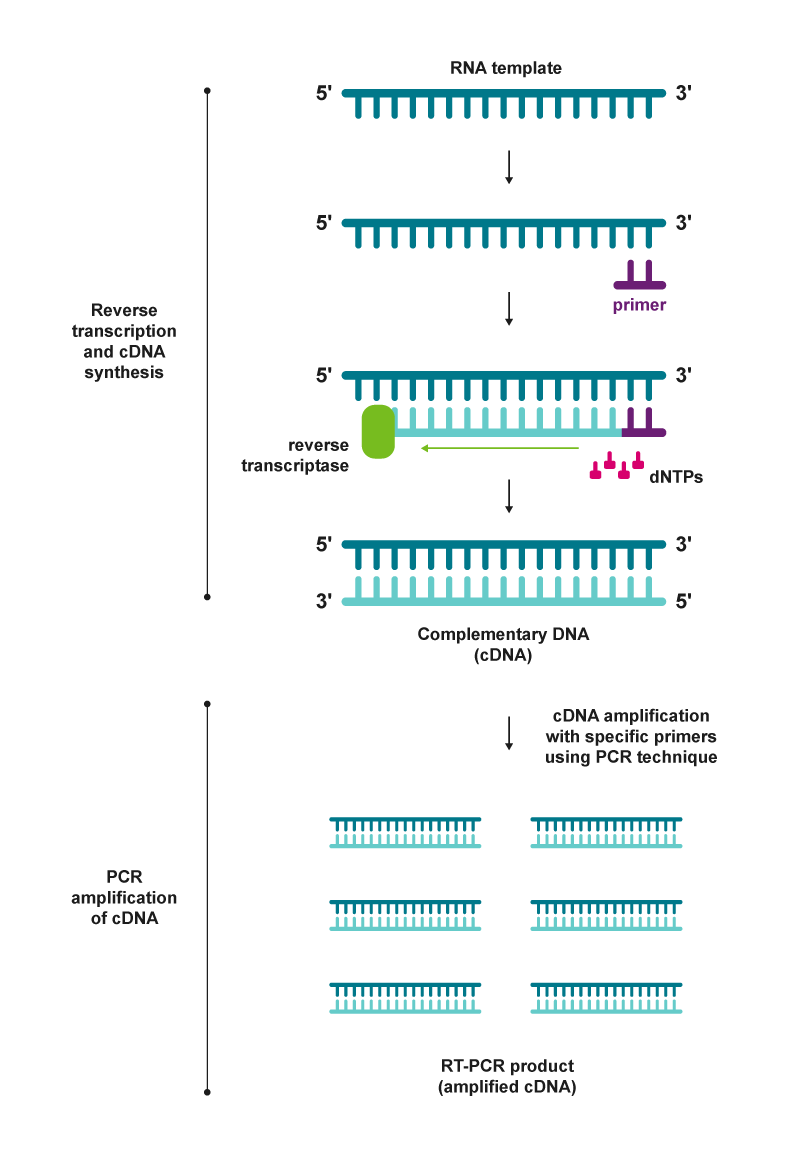
|
| Figure 2: RT-PCR steps |
The RT-PCR reaction can be carried out as a one-step or a two-step process. In one-step RT-PCR, the cDNA synthesis and amplification are conducted in a single reaction tube, whereas in two-step RT-PCR, two separate reaction tubes are used.5 The advantages, disadvantages and application considerations of each method are highlighted in figure 3.
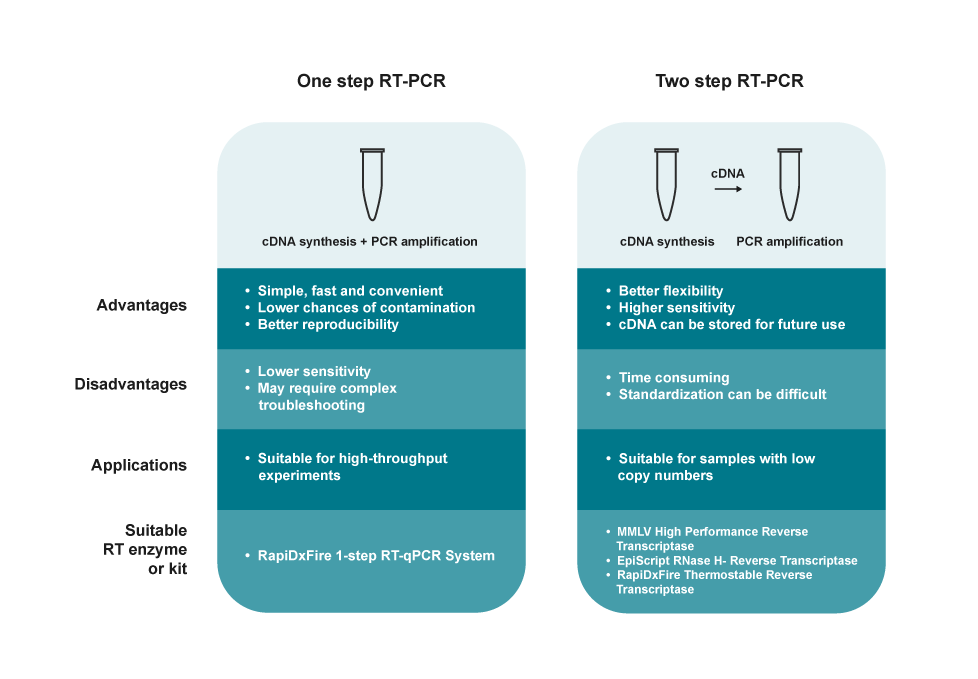 |
| Figure 3: One-step vs. two-step RT-PCR |
Next-generation sequencing (NGS)
NGS is an ultra-high-throughput sequencing method that enables large-scale sequencing of DNA or RNA from tissue/cell samples. Over the last decade, it has become a widely popular technique and has revolutionised genomics and transcriptomics.6
RNA-sequencing (RNA-seq) using NGS platforms can provide a comprehensive overview of the RNA transcriptome in a cell. It gives valuable insights into mRNA expression levels, alternate splicing, allelic expression, and the regulatory functions of different types of non-coding (nc) RNAs, such as pre-mRNA, micro-RNA and long ncRNAs. Whole-exome sequencing or targeted transcriptome sequencing are widely used for drug discovery research and translational studies. The application of RNA-seq in clinical diagnostics is also gaining momentum.7
The main steps in an RNA-seq workflow are RNA extraction, cDNA conversion, sequencing library preparation and NGS sequencing (figure 4). Success in RNA-sequencing relies on the selection and efficiency of the reverse transcriptase enzyme used for NGS cDNA preparation. Ideally, the reverse transcriptase must be thermostable and efficient to capture low-abundance and full-length RNA sequences (Check out the EpiScript RNase H- Reverse Transcriptase Kit).
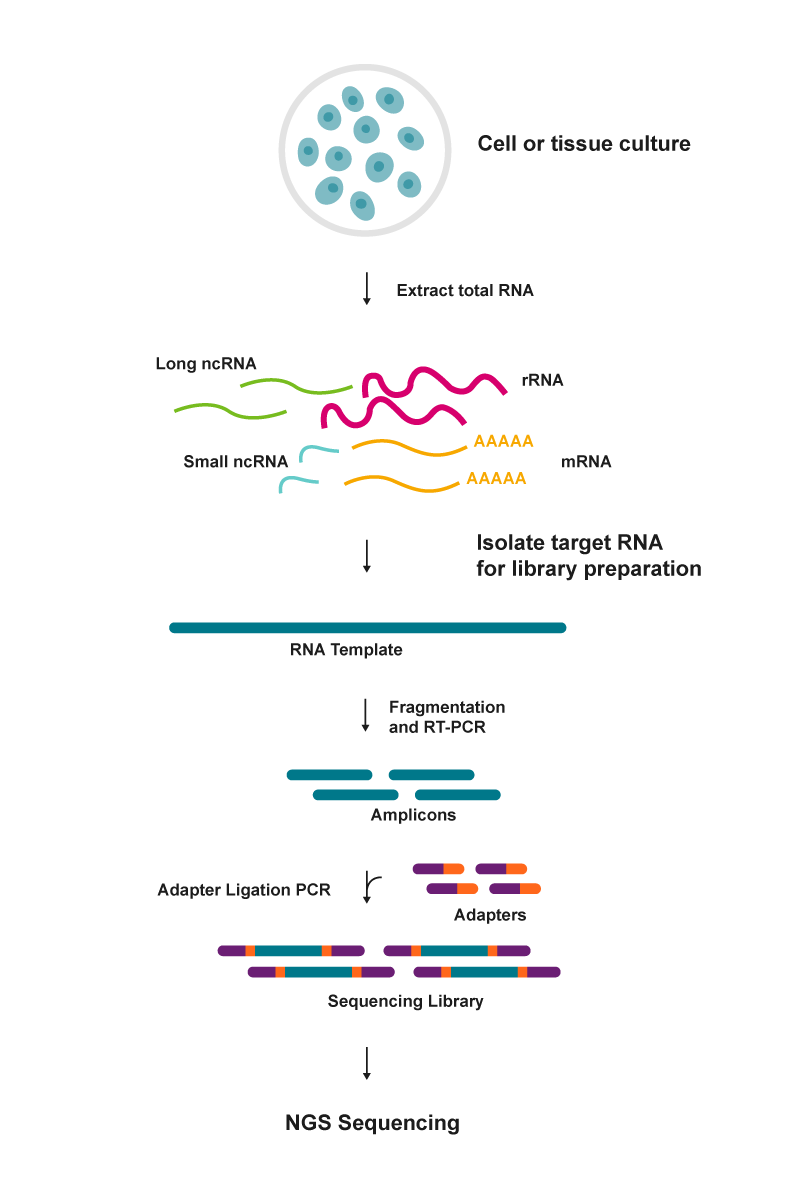
|
| Figure 4: Basic steps in an RNA sequencing workflow |
Use of reverse transcriptases in novel techniques
Prime editing
This is a novel ‘search-and-replace’ genome editing technology with almost any type of genome rewriting capacity. It is a CRISPR/Cas9- based technology where a mutant Cas9 nickase is fused to a reverse transcriptase and a prime editing guide RNA (pegRNA).8
Epitranscriptomics
Epitranscriptomics is a new field of research on RNA modifications and the regulation of non-coding RNAs, and their role in disease pathologies.9 The advent of high-throughput RNA-seq methods has enabled the identification of millions of transcriptome-wide RNA modifications. Epitranscriptomics employs highly efficient reverse transcriptases that can capture full-length RNA sequences and RNAs with complex secondary structures.
Find the best reverse transcriptase enzyme for your experiments
LGC Biosearch Technologies™ offers a range of reverse transcriptase enzymes suitable for various molecular biology experiments. We also partner with scientists to provide customisation solutions to suit your assay requirements and applications.
References
- Sarafianos SG, Marchand B, Das K, et al. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. Journal of molecular biology, 385(3), 693–713. https://doi.org/10.1016/j.jmb.2008.10.071 Published 2009. Accessed October 10, 2022
- Goff SP. Retroviral Reverse Transcriptase: Synthesis, Structure, and Function. Journal of acquired immune deficiency syndromes, 3(8), 817–831. https://pubmed.ncbi.nlm.nih.gov/1694894/. Published 1990. Accessed October 10, 2022
- Gerard G, Potter RJ, Smith MD, Rosenthal K et al. The role of template-primer in protection of reverse transcriptase from thermal inactivation. Nucleic acids research, 30(14), 3118–3129. https://doi.org/10.1093/nar/gkf417. Published 2002. Accessed October 11, 2022
- Ying SY. Complementary DNA libraries. Molecular biotechnology, 27(3), 245–252. https://doi.org/10.1385/MB:27:3:245. Published 2004. Accessed October 11, 2022
- Wacker MJ, Godard MP. Analysis of One-Step and Two-Step Real-Time RT-PCR Using SuperScript III. Journal of Biomolecular Techniques : JBT, 16(3), 266-271. https://doi.org/https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2291734/. Published 2005. Accessed October 11, 2022
- Kukurba KR, Montgomery SB. RNA Sequencing and Analysis. Cold Spring Harbor protocols, 2015(11), 951–969. https://doi.org/10.1101/pdb.top084970. Published 2015. Accessed October 11, 2022
- Byron S, Van Keuren-Jensen K, Engelthaler D. et al. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet 17, 257–271 (2016). https://doi.org/10.1038/nrg.2016.10. Published 2016. Accessed October 12, 2022
- Martín-Alonso S, Frutos-Beltrán E, Menéndez-Arias L. Reverse Transcriptase: From Transcriptomics to Genome Editing. Trends in biotechnology, 39(2), 194–210. https://doi.org/10.1016/j.tibtech.2020.06.008. Published 2021. Accessed October 12, 2022
- Song H, Liu D, Dong S, et al. Epitranscriptomics and epiproteomics in cancer drug resistance: therapeutic implications. Signal transduction and targeted therapy, 5(1), 193. https://doi.org/10.1038/s41392-020-00300-w. Published 2020. Accessed October 12, 2022





