Originally published : Thu, January 22, 2015 @ 3:28 PM
Updated : Mon, November 16, 2015 @ 5:30 PM
In a webinar linked below, Dr. James Willey describes a two colour fluorometric assay for more robust quantitation of nucleic acids in highly degraded FFPE and FNA samples. Willey’s method, described in an article published in the journal PLoS One, employs a patented mixture of internal and external standards paired with a pre-amplification step. This system enables measurement of transcription levels of three lung cancer-associated genes and a control gene in clinical samples. All of the fluorometric probes used in the method are labeled with Black Hole Quencher® dyes in BHQplus® probes. The compact design of BHQplus probes is optimal for this application as it confers enhanced sequence discrimination for each target. The webinar takes the audience through assay development, reagent preparation, and the analytical validation experiments.
Challenging samples: fine needle aspirate (FNA) and FFPE specimens
Formalin-fixed paraffin-embedded (FFPE) specimens have long been the preferred sample of choice for preservation of clinical specimens destined for long-term, room temperature storage. This preservation method was originally developed to maintain tissue morphology for histological analysis in a time when there was little consideration of nucleic acid preservation. Despite the limitations of FFPE sample preparation, these samples are still important to clinical diagnostics and pathology studies. Millions of specimens are archived in longitudinal biopsy collections with known clinical outcomes. Retrospective analysis of these FFPE specimens with modern genetic methods presents a dramatic opportunity for biomarker discovery, pairing nucleic acid analysis with disease association. Fine Needle Aspirate (FNA) biopsy cell blocks are being used with increasing frequency in the diagnosis of cancer and inflammatory conditions. FNA cell blocks, frequently formalin-fixed and paraffin embedded for ease of cytomorphologic evaluation, are now being probed with molecular diagnostics.
Nucleic acid integrity within FFPE specimens
Banked FFPE specimens present numerous analytical challenges due to harsh fixation methods which create sporadic degradation of nucleic acid integrity. Further complicating the use of material from these samples is the non-uniformity of not only their preservation, but also the extraction of nucleic acids. Sample storage methods and preparation can vary widely according to fixation protocol, length of storage, ambient conditions, and specimen size.
Measuring transcript abundance in diagnostics
Historically, in order to quantify results obtained by real-time qRT-PCR, there have been generally two methods used. The first and preferred method uses a standard curve created by a dilution series of known concentrations of template. A common alternative method of quantification compares the cycle threshold (Ct) value of the sample to the Ct value of a control, such as a non-treated sample or reference calibrator, followed by normalisation of the Ct values of the calibrator and sample to an appropriate endogenous housekeeping gene.
In the webinar (see below) Dr. Willey explains a new normalisation method employing synthetic competitive internal standards that have been used for single analytes in FDA-approved diagnostics such as the Roche COBAS® Ampliscreen HIV-1 Test. With advances in personalised medicine, there is increased interest in the measurement of transcript abundance of multiple genes using qRT-PCR data as a prognostic of disease progression allowing selection of the most efficacious treatment for an individual's cancer.
New assay format eliminates the need for an external standard curve
Dr. Willey described the competitive qRT-PCR assay, designed using a multiplex pre-amplification reaction with a mix of synthetic internal standards (MIS), followed by a second round of amplification using BHQplus probes for detection and quantification of a gene of interest. This assay does not require an external standard curve. The MIS also controls for interfering substances and false negatives. This approach to expression profiling allows an efficient method of creating normalised data in difficult samples using the unique short probe binding properties of BHQplus probes for challenging targeted sites.
Lung cancer diagnostic test (LCDT)
The assay they developed makes use of a sequence-specific FAM-labeled probe homologous for the native template (NT). A second sequence-specific Quasar® dye-labeled probe is designed for the internal standard (IS), with a 4-6 base difference from the native template (NT). The first round of PCR allows conservation of sample and pre-amplification of the targets, while the second round of PCR uses a single primer pair and competitive NT and IS BHQplus probes for each gene of interest. The NT and IS probes compete for the same primers and amplify with the same efficiency. The mixture of synthetic internal standards (MIS) for each of four genes controls for interfering substances in the reaction, pipetting variation, and prevents false negatives.
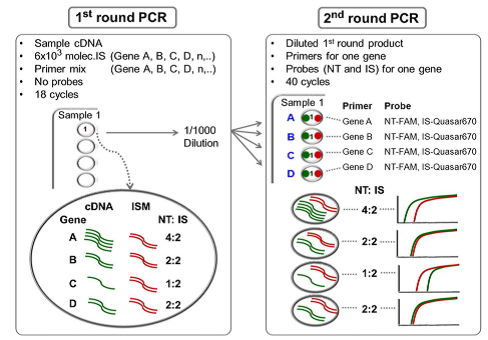
Dr. Willey provides a detailed description of the two colour fluorometric assay development: preparation of synthetic internal standards for each gene, mixture of internal standards (MIS), analytical validation, and an external standard mixture (ESM) for inter-experimental variation. A detailed outline and data are provided for a four-gene lung cancer diagnostic test (LCDT) used to analyse trans-thoracic FNA samples.
View the webinar
Dr. James Willey is a co-founder, inventor and medical advisor for Accugenomics, and the George Isaac Professor for Cancer Research, University of Toledo Health Sciences Campus. The technology described, published in PLoS ONE1 is the subject of US patents (e.g., U.S. 7,527,930), licensed to Accugenomics, Inc.
References:
1. Yeo J, Crawford EL, Blomquist TM, Stanoszek LM, Dannemiller RE, et al. (2014) A Multiplex Two-Color Real-Time PCR Method for Quality-Controlled Molecular Diagnostic Testing of FFPE Samples. PLoS ONE 9(2): e89395. doi:10.1371/journal.pone.0089395

