Originally published : Tue, April 23, 2013 @ 7:02 PM
Updated : Mon, September 19, 2022 @ 2:10 PM
A "Referral from the Doctor" Blog Article-
Quantitative reverse transcription polymerase chain reaction (RT-qPCR) is a sensitive and
robust method for analyzing RNA. It has been used to corroborate the findings of other genetic methods, such as microarray and RNAse protection assays, and has become the gold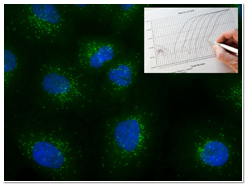 standard for quantifying changes in gene expression. Stellaris FISH is a new RNA detection method that provides direct quantification of transcripts in situ, with potential to become the standard for mRNA and lncRNA interrogation using microscopy.
standard for quantifying changes in gene expression. Stellaris FISH is a new RNA detection method that provides direct quantification of transcripts in situ, with potential to become the standard for mRNA and lncRNA interrogation using microscopy.
Both RT-qPCR and Stellaris FISH probes use a fluorescent signal as a means of detection. Gene expression analysis using RT-qPCR is an in vitro reaction involving extracted RNA, with an increasing fluorescent signal indicating exponential amplification of a target sequence. Accumulated signal is plotted in relation to the number of thermal cycles to cross an arbitrary threshold, or cycles to quantification (Cq). The Cq values are correlated to copy numbers assigned by a standard curve, which can then be normalized to a reference gene. In contrast, the Stellaris FISH method does not use signal amplification but instead interrogates individual RNA targets through the direct binding of up to 48 fluorescent dye-labeled probes in situ. The fluorescent signal localizes to the site of each transcript, and can be visualized within the specimen as discreet spots that are counted to achieve a digital output. Stellaris FISH probes are perfect for localizing the transcripts to subcellular compartments or across a heterogeneous tissue sample. While qPCR destroys the morphology of the sample during RNA extraction, this technique remains well-suited to screen a large input sample for a rare transcript.
Quantification and variance
Quantification by RT-qPCR is made complicated by analysis of reaction kinetics, and has inherent variability due to its indirect detection method. Sequential steps of nucleic acid extraction, reverse transcription, and PCR amplification each have a finite efficiency, and may vary from one preparation to the next. QPCR usually outputs an average from a population of cells, overlooking the impact of individual cells with extreme copy number variation. RT-qPCR has statistical power, by virtue of the sheer number of cells analyzed per sample, and established methods of quantification that matured in parallel with the technique. In contrast, quantification by Stellaris is straightforward due to its direct detection method, requiring no additional biochemistry beyond sample fixation and oligo hybridization. It provides granular visualization of the transcripts at a sub-cellular level, enabling powerful investigations into localization as a form of regulation. However, automated methods of quantification are still being formalized, requiring advanced algorithms to process microscope images, recognize cellular boundaries, and assign a signal threshold to better demarcate the true transcripts from background artifacts.
Stellaris FISH complements RT-qPCR findings
RT-qPCR and Stellaris methods both provide measures of gene expression through relative or absolute quantification, but excel in different applications. Combined, they provide complementary information for particularly rigorous studies. For example, qPCR-based screening might uncover a signature pattern of gene expression from within a tumor biopsy. The individual biomarkers of interest can then be further characterized with Stellaris FISH to image their discreet localization across the tissue section, and even differentiate stem-like cancer cells from within the surrounding tumor.
Written by: Christina Ferrell, Ph.D.
References:
Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008; 5(10):877–879
Stochastic mRNA Synthesis in Mammalian Cells. PLoS Biol. 2006; 4(10):e309
Lysogen stability is determined by the frequency of activity bursts from the fate-determining gene. Mol Syst Biol. 2010 Nov 30; 6:440.
General properties of transcriptional time series in Escherichia coli. Nat Genet. 2011 Jun; 43(6):554-60.
Cell-to-cell variability of alternative RNA splicing. Mol Syst Biol. 2011 Jul 5; 7:506.
Single-Molecule Approaches to Stochastic Gene Expression. Annu Rev Biophys. 2009; 38:255–270

