Originally published : Wed, November 18, 2009 @ 4:01 PM
Updated : Mon, September 19, 2022 @ 2:10 PM
A "Referral from the Doctor" Blog Article-
Competitive qRT-PCR has become a lost art. Early titans of probe-based PCR used this technique to minimize variability between samples and experiments due to differences in reverse transcription and amplification efficiency. This technique facilitates the absolute measurement of initial gene target copy number based on known copy numbers of a control template.
This is accomplished through the addition of an internal control, a competitor sequence to the mRNA target in the sample. The competitor sequence is similar enough to use the same primer pairs as the target sequence but has a unique central sequence allowing for analysis with a different probe and reporter. Plasmid DNA or in-vitro transcribed RNA is commonly used as a competitor sequence and standard for quantification. To detect the initial copy number of a gene of interest, multiple replicates of a fixed amount of the target mRNA are dispensed into tubes. In the same tubes, 10 fold serial dilutions of competitor RNA are "spiked" in sequentially. Using one set of primers and two fluorogenic probes labeled with different dyes, real-time PCR data can be collected for both sequences simultaneously.
To determine copy number, the mean Cycles to Threshold (CT) values from the target sequence and the mean CT values of the competitor serial dilutions are plotted on the y axis, against the known copy number of the competitor sequence on the x axis. The number of initial copies of the target gene can be calculated from the theoretical equivalence point, where the CT of the target gene equals the CT of the competitor sequence - the point of intersection for each linear plot. It is essential that the efficiency of both amplifications be greater than 88% and furthermore within 5% of each other for accurate analysis.
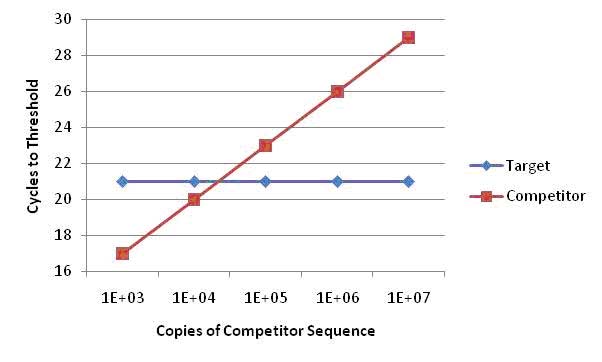
This method of quantitative PCR is returning to the forefront of gene expression analysis, particularly for low expressing genes or those difficult to amplify. Success in the modern age is dependent upon the equipment capability, the stringency of validation and selection of fluorogenic probes. For a catalog of available probe formats, visit: http://www.biosearchtech.com/products/fluorogenic-probes-and-primers
Thus, we can rise above and see beyond because we stand on the shoulders of giants.
-concept attributed to Bernard of Chartres
Written by: Christina Ferrell, Ph.D., Technical Applications Specialist
References:
H. Tani, T. Kanagawa, S. Kurata, T. Teramura, K. Nakamura, S. Tsuneda, N. Noda. Quantitative method for specific nucleic acid sequences using competitive polymerase chain reaction with an alternately binding probe. Anal. Chem. (2007) 79(3):974-979
E. Barbieri, G. Riccioni, A. Pisano, D. Sisti, S. Zeppa, D. Agostini, V. Stocchi. Competitive PCR for quantitation of a cytophaga-flexibacter-bacteroides phylum bacterium associated with the tuber borchiivittad. Mycelium. Applied and Environmental Microbiology (2002)68(12):6421-6424
C. Orlando, P. Pinzani, M. Pazzagli. Developments in Quantitative PCR.ClinChem Lab Med (1998) 36(5):255-269

