Originally published : Tue, February 23, 2010 @ 10:06 PM
 Updated : Thu, March 23, 2023 @ 11:57 AM
Updated : Thu, March 23, 2023 @ 11:57 AM
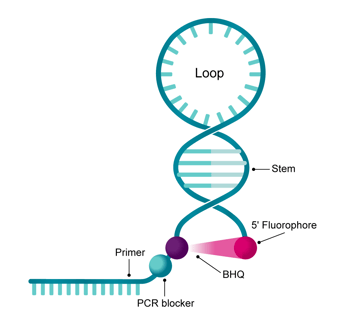
Scorpions™ Primers are dual-labeled FRET probes that combine a Molecular Beacon-like probe structure and a PCR primer element in a single oligonucleotide, allowing for specific target amplification and advanced target detection through a unimolecular or single oligonucleotide driven mechanism.
Structure
What parts are similar to dual-labeled Black Hole Quencher™ (BHQ) probes?
A BHQ dye and a 5'-fluorophore covalently bound to the oligonucleotide termini.
What parts are similar to Molecular Beacons?
A target-binding region, comprised of a stem-loop structure resembling a "closed hairpin".
What parts are unique?
• A 3' template-binding region representing the primer region;
• A PCR blocker, often a hexethylene glycol modification linker sequence and;
• An internal thymidine labeled with a BHQ.
What is its native conformation at melting and annealing temperatures?
Melted - random coil conformation
Annealed (perfect match) - stable double helix, probe-target hybrid (high signal to noise ratio)
Annealed (mismatch) - hairpin conformation (FRET quenched, no signal).
Mechanism
How does it yield signal?
The target-binding region is located within the loop portion of the hairpin conformation and sets atop an annealed stem region. When a perfectly matched complementary sequence is available, the probe region hybridises to the complementary sequence. Conformational changes associated with hybridisation force the hairpin stem region open, separating the fluorophore and quencher, decreasing FRET quenching and releasing fluorescence.
How does it amplify its own target?
During the first round of amplification, the template-binding region hybridises to the complementary sequence of the template DNA and extends to form a primer extension product. The primer extension product contains the desired target sequence. The probe region remains in a closed hairpin conformation until the next cycle of amplification. The presence of a PCR blocker in the linker sequence prevents the polymerase-mediated duplication of the probe region.
How does it detect the target it has amplified?
During the second round of amplification, the target-binding region of the probe sequence hybridises to the complementary sequence within the primer extension product. Hybridisation forces the hairpin open, releasing fluorescence.
What happens with each consecutive PCR cycle?
The primer extension product and connected probe region, establishes a temperature-dependent equilibrium between the hybridised conformation and the random coil conformation. Therefore, each Scorpions Primer accounts for only one product amplification per experiment.
The unimolecular mechanism allows for the linear amplification of targets with initially high copy number. For the quantitative analysis of low copy number sequences, it is essential that the reverse primer be included in the PCR master mix to enable exponential amplification of the target sequence.
What if the available target is a mismatch?
Mismatched hybrids are less stable than reformation of the hairpin stem. Therefore signal is only produced when the target binding region hybridises to a perfect-match target sequence. This enables specificity of signal generation to be accurate at the level of single nucleotide polymorphisms (SNP).
Advantages of Scorpions Primers
- Rapid hybridisation - the proximity of the probe region and the target sequence kinetically favours formation of the probe-template hybrid over template duplex re-annealing. The unimolecular event enables rapid signal generation during hybridisation.
- High signal to noise ratio - the efficiency of FRET quenching, when in the unhybridised state, decreases errant fluorescence and enables a high signal to noise ratio with hybridisation.
- High specificity - single base mismatches can be detected.
- Post-PCR melt curve analyses - Scorpions Primers do not require the enzymatic activity of a polymerase for fluorescent signal generation in qPCR and under non-hydrolytic conditions allow for post-PCR melt curve analyses.
Applications
Written by: Christina Ferrell, Ph.D., Technical Applications Specialist
- Whitcombe, D.M., Theaker, J., Gibson, N.J., Little, S. "Methods for detecting target nucleic acid sequences". United States Patent 6270967. Aug. 07, 2001
- Whitcombe, D., Theaker, J., Guy, S.P., Brown, T., Little, S. "Detection of PCR products using self-probing amplicons and fluorescence". Nature Biotechnology 17 (1999): 804-807.
- Thelwell, N., Millington, S., Solinas, A., Booth, J., Brown, T. "Mode of Action and application of Scorpion primers to mutation detection". Nucleic Acids Research 28(19) (2000): 3752-3761.
- Solinas, A., Brown, L.J., McKeen, C., Mellor, J.M., Nicol, J.T.G., Thelwell, N., Brown, T. "Duplex Scorpion primers in SNP analysis and FRET applications". Nucleic Acids Research 29(20) (2001): e96.
- Bustin, S.A. A-Z of Quantitative PCR (IUL Biotechnology Series). La Jolla, California: International University Line, 2004.


