Originally published : Mon, August 22, 2016 @ 8:22 PM
Updated : Wed, March 29, 2023 @ 11:04 AM
As the Zika virus outbreak spreads, a wave of international concern has grown, and scientists have scrambled to overcome the significant challenge of diagnosing the virus so it can be treated effectively. Fortunately, a new multiplexed-qPCR assay using Dual-Labeled BHQ™ Probes from LGC Biosearch Technologies has recently been developed that can distinguish between Zika and other viral infections that cause similar symptoms.
Zika is a single-stranded RNA arbovirus, spread by the Aedes aegypti and Ae. albopictus mosquitoes1. These mosquito species are also the same vectors that spread other viral pathogens such as Chikungunya and Dengue, both of which can present similar symptoms as those of Zika. It’s been a true clinical challenge to diagnose and differentiate between Chikungunya, Dengue, and Zika infections.
On February 1, 2016 the World Health Organization (WHO) released a statement of public health concerns over a link between Zika virus and prevalence of microcephaly in the offspring of infected individuals2. Of all the South American countries affected by Zika, Brazil has been hardest hit, with an estimated 1.5 million cases since 20153. Subsequent to the announcement, concern has grown internationally due to the 2016 Olympic games in Rio de Janeiro, Brazil, coinciding with the outbreak. Recently, Centres for Disease Control and Prevention (CDC) issued a historic travel advisory relating to the continental United States due to localized transmission within a region of Miami, FL4,5. Zika cases have also been reported in numerous other US locations, particularly in the territory of Puerto Rico which is under a public health emergency as of August 12, 2016 (see inset map). CDC advises travellers to take precautions against Zika, and recommends those experiencing symptoms to meet with their physicians6.
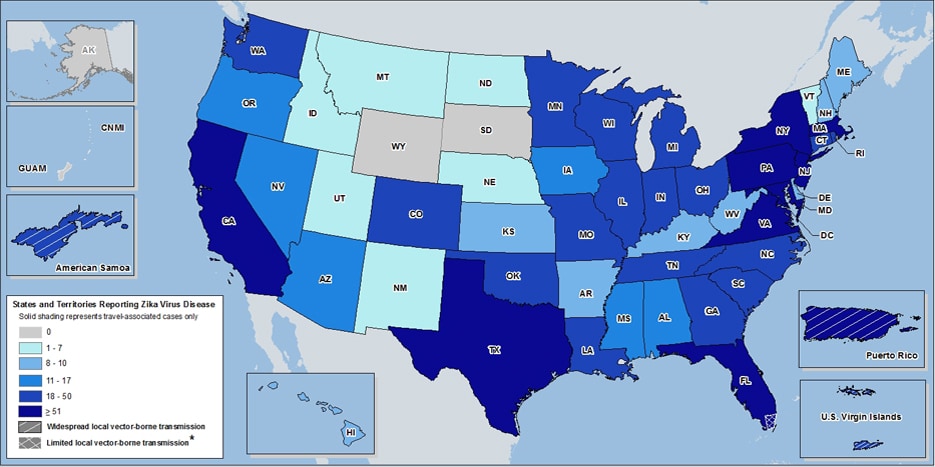
Heat map of ArboNET reported Zika cases in US as of August 17th, 2016. Map source: CDC;NCEZID;DVBD, Accessed 8/23/16.
Current tests for Zika virus infection include serological methods such as ELISA (enzyme-linked immunosorbent assay) and PRNT (plaque-reduction neutralization tests)7. These methods are generally used for diagnosis during the convalescent phase of the infection8,9. However, the CDC also describe molecular methods for detection of Zika virus during the acute phase of infection, through RT-qPCR (quantitative reverse transcription PCR)10.

The upsurge of Zika infection poses an immediate challenge to incorporate a Zika test into existing assays and protocols, which were oriented toward Chikungunya and Dengue prior to the recent outbreak11-14. Typically each test is performed as an individual reaction upon a single sample or specimen, with established protocols using multiple separate tests for each of the three viruses11,15,16. Recently, a team of scientists from Stanford University, Sustainable Sciences Institute (Nicaragua), Nicaragua Ministry of Health, and University of California, Berkeley published a study titled “Single-Reaction Multiplex Reverse Transcription PCR for Detection of Zika, Chikungunya, and Dengue Viruses”. This publication describes a multiplex method called the ZCD assay, to consolidate all three tests into a single multiplex reaction, and determine whether Zika, Chikungunya, and/or Dengue virus are each present within the sample under investigation.
The ZCD assay developed by this group uses custom Dual-Labeled BHQ™ Probes from LGC Biosearch Technologies to detect signature nucleic acid sequences from the viruses independently while amplifying them all simultaneously. In the study the authors indicate that the newly developed ZCD assay showed improved detection of Zika virus compared to previously published assays11, and similar sensitivity while detecting Chikungunya and Dengue viruses. The authors also assert that development of multiplexed assays can potentially streamline the laboratory workflow and reduce the costs by conserving sample materials.
With the recent end of the Olympics, it is expected that as many as 500,000 people worldwide could have travelled to Brazil to watch the games17. The large influx of visitors to the continent has public health agencies keeping a watchful eye on the situation. Fortunately, there are scientific methods and technologies to enable rapid epidemiological assessment, equipping public health workers with the surveillance tools to combat the spread of infectious disease.
References:
- https://www.cdc.gov/zika/about/questions.html Accessed: 08/23/2016
- WHO statement on the first meeting of the International Health Regulations (2005) (IHR 2005) Emergency Committee on Zika virus and observed increase in neurological disorders and neonatal malformations Accessed: 08/23/2016
- http://www.who.int/emergencies/zika-virus/situation-report/who-zika-situation-report-12-02-2016.pdf Accessed: 08/23/2016
- CDC issues travel guidance related to Miami neighborhood with active Zika spread Accessed: 08/23/2016
- Advice for people living in or traveling to South Florida Accessed: 08/23/2016
- http://www.cdc.gov/zika/symptoms/symptoms.html Accessed: 08/23/2016
- Interim Guidance for Interpretation of Zika Virus Antibody Test Results. Accessed: 08/23/2016
- Laboratory testing for Zika virus infection. Accessed: 08/23/2016
- Zika virus (ZIKV) Surveillance in the Americas: Recommendations for laboratory detection and diagnosis; 2016. Accessed: 08/23/2016
- http://www.cdc.gov/zika/hc-providers/types-of-tests.html Accessed: 08/23/2016
- Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis. 2008 Aug;14(8):1232-9. doi: 10.3201/eid1408.080287
- Faye O, Faye O, Dupressoir A, Weidmann M, Ndiaye M, Alpha Sall A. One-step RT-PCR for detection of Zika virus. J Clin Virol. 2008;43:96–101. 10.1016/j.jcv.2008.05.005
- Waggoner JJ, Pinsky BA. Zika virus: diagnostics for an emerging pandemic threat. J Clin Microbiol. 2016;54:860–7.;54:860–7.
- Waggoner JJ, Gresh L, Mohamed-Hadley A, Ballesteros G, Davila MJ, Tellez Y, Sahoo MK, Balmaseda A, Harris E, Pinsky BA. Single-reaction multiplex reverse transcription pcr for detection of zika, Chikungunya, and dengue viruses. Emerg Infect Dis. 2016;22(7):1295–1297. doi: 10.3201/eid2207.160326.
- Lanciotti RS, Kosoy OL, Laven JJ, Panella AJ, Velez JO, Lambert AJ, et al. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis. 2007;13:764–7. 10.3201/eid1305.070015
- Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase–polymerase chain reaction. J Clin Microbiol. 1992;30:545–51
- Simon Romero and Rebecca R. Ruiz. "Experts Weigh the Risks of Zika Spreading Via Rio Olympics." New York Times, (2016): http://www.nytimes.com/2016/01/29/world/americas/brazil-zika-rio-olympics.html. Accessed: 08/23/2016

